Insight into the autoproteolysis mechanism of the RsgI9 anti-sigma factor from Clostridium thermocellum.
Takayesu, A., Mahoney, B.J., Goring, A.K., Jessup, T., Ogorzalek Loo, R.R., Loo, J.A., Clubb, R.T.(2024) Proteins 92: 946-958
- PubMed: 38597224
- DOI: https://doi.org/10.1002/prot.26690
- Primary Citation of Related Structures:
8U9O - PubMed Abstract:
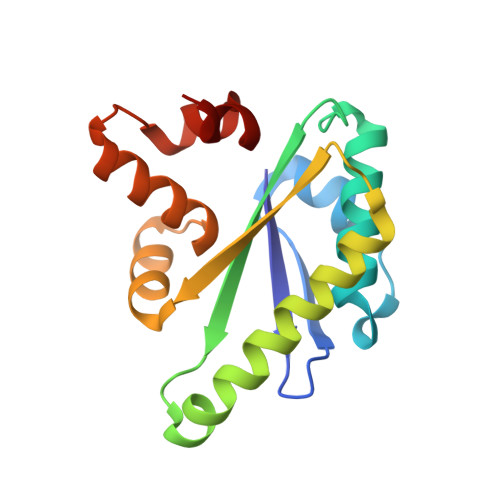
Clostridium thermocellum is a potential microbial platform to convert abundant plant biomass to biofuels and other renewable chemicals. It efficiently degrades lignocellulosic biomass using a surface displayed cellulosome, a megadalton sized multienzyme containing complex. The enzymatic composition and architecture of the cellulosome is controlled by several transmembrane biomass-sensing RsgI-type anti-σ factors. Recent studies suggest that these factors transduce signals from the cell surface via a conserved RsgI extracellular (CRE) domain (also called a periplasmic domain) that undergoes autoproteolysis through an incompletely understood mechanism. Here we report the structure of the autoproteolyzed CRE domain from the C. thermocellum RsgI9 anti-σ factor, revealing that the cleaved fragments forming this domain associate to form a stable α/β/α sandwich fold. Based on AlphaFold2 modeling, molecular dynamics simulations, and tandem mass spectrometry, we propose that a conserved Asn-Pro bond in RsgI9 autoproteolyzes via a succinimide intermediate whose formation is promoted by a conserved hydrogen bond network holding the scissile peptide bond in a strained conformation. As other RsgI anti-σ factors share sequence homology to RsgI9, they likely autoproteolyze through a similar mechanism.
- Department of Chemistry and Biochemistry, University of California, Los Angeles, Los Angeles, California, USA.
Organizational Affiliation: