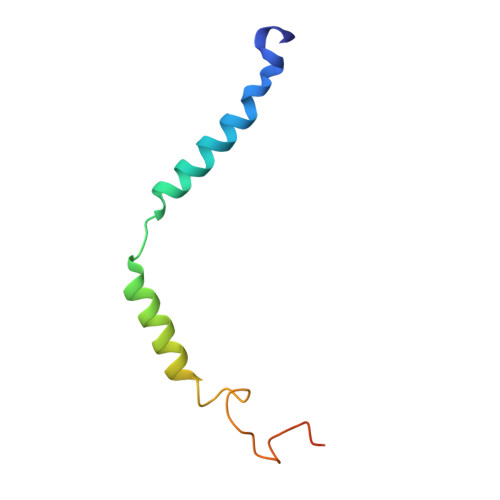
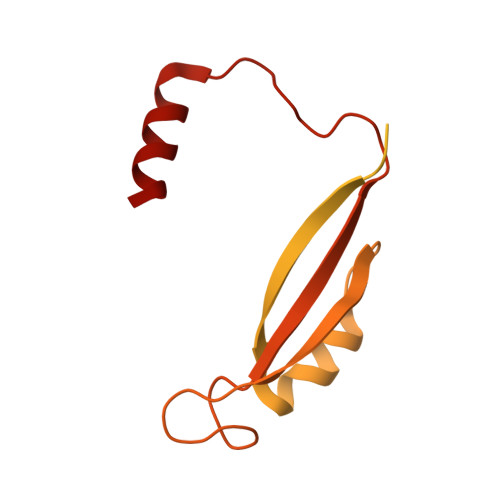
Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Nguyen, D.M., Rath, D.H., Devost, D., Petrin, D., Rizk, R., Ji, A.X., Narayanan, N., Yong, D., Zhai, A., Kuntz, D.A., Mian, M.U.Q., Pomroy, N.C., Keszei, A.F.A., Benlekbir, S., Mazhab-Jafari, M.T., Rubinstein, J.L., Hebert, T.E., Prive, G.G.(2024) Proc Natl Acad Sci U S A 121: e2315018121-e2315018121
- PubMed: 38625940
- DOI: https://doi.org/10.1073/pnas.2315018121
- Primary Citation of Related Structures:
8U7Z, 8U80, 8U81, 8U82, 8U83, 8U84 - PubMed Abstract:
Heterotrimeric G proteins can be regulated by posttranslational modifications, including ubiquitylation. KCTD5, a pentameric substrate receptor protein consisting of an N-terminal BTB domain and a C-terminal domain, engages CUL3 to form the central scaffold of a cullin-RING E3 ligase complex (CRL3 KCTD5 ) that ubiquitylates Gβγ and reduces Gβγ protein levels in cells. The cryo-EM structure of a 5:5:5 KCTD5/CUL3 NTD /Gβ 1 γ 2 assembly reveals a highly dynamic complex with rotations of over 60° between the KCTD5 BTB /CUL3 NTD and KCTD5 CTD /Gβγ moieties of the structure. CRL3 KCTD5 engages the E3 ligase ARIH1 to ubiquitylate Gβγ in an E3-E3 superassembly, and extension of the structure to include full-length CUL3 with RBX1 and an ARIH1~ubiquitin conjugate reveals that some conformational states position the ARIH1~ubiquitin thioester bond to within 10 Å of lysine-23 of Gβ and likely represent priming complexes. Most previously described CRL/substrate structures have consisted of monovalent complexes and have involved flexible peptide substrates. The structure of the KCTD5/CUL3 NTD /Gβγ complex shows that the oligomerization of a substrate receptor can generate a polyvalent E3 ligase complex and that the internal dynamics of the substrate receptor can position a structured target for ubiquitylation in a CRL3 complex.
- Princess Margaret Cancer Centre, University Health Network, Toronto, ON M5G 1L7, Canada.
Organizational Affiliation: