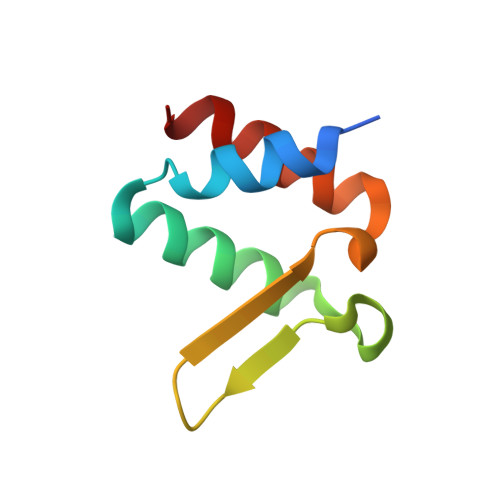
Molecular insight into interactions between the Taf14, Yng1 and Sas3 subunits of the NuA3 complex.
Nguyen, M.C., Rostamian, H., Raman, A., Wei, P., Becht, D.C., Erbse, A.H., Klein, B.J., Gilbert, T.M., Zhang, G., Blanco, M.A., Strahl, B.D., Taverna, S.D., Kutateladze, T.G.(2024) Nat Commun 15: 5335-5335
- PubMed: 38914563
- DOI: https://doi.org/10.1038/s41467-024-49730-y
- Primary Citation of Related Structures:
8U77 - PubMed Abstract:
The NuA3 complex is a major regulator of gene transcription and the cell cycle in yeast. Five core subunits are required for complex assembly and function, but it remains unclear how these subunits interact to form the complex. Here, we report that the Taf14 subunit of the NuA3 complex binds to two other subunits of the complex, Yng1 and Sas3, and describe the molecular mechanism by which the extra-terminal domain of Taf14 recognizes the conserved motif present in Yng1 and Sas3. Structural, biochemical, and mutational analyses show that two motifs are sandwiched between the two extra-terminal domains of Taf14. The head-to-toe dimeric complex enhances the DNA binding activity of Taf14, and the formation of the hetero-dimer involving the motifs of Yng1 and Sas3 is driven by sequence complementarity. In vivo assays in yeast demonstrate that the interactions of Taf14 with both Sas3 and Yng1 are required for proper function of the NuA3 complex in gene transcription and DNA repair. Our findings suggest a potential basis for the assembly of three core subunits of the NuA3 complex, Taf14, Yng1 and Sas3.
Organizational Affiliation:
Department of Pharmacology, University of Colorado School of Medicine, Aurora, CO, 80045, USA.