A role for the ATP-dependent DNA ligase lig E of Neisseria gonorrhoeae in biofilm formation.
Pan, J., Singh, A., Hanning, K., Hicks, J., Williamson, A.(2024) BMC Microbiol 24: 29-29
- PubMed: 38245708
- DOI: https://doi.org/10.1186/s12866-024-03193-9
- Primary Citation of Related Structures:
8U6X - PubMed Abstract:
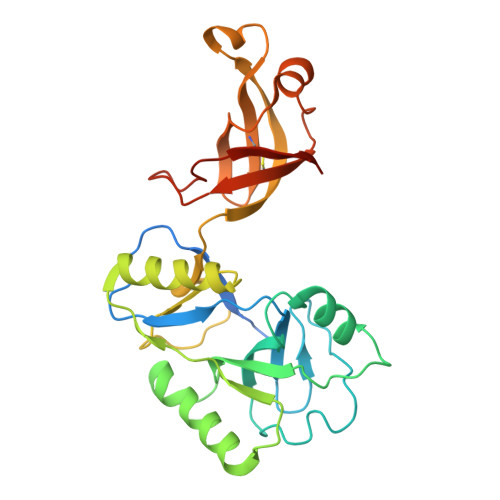
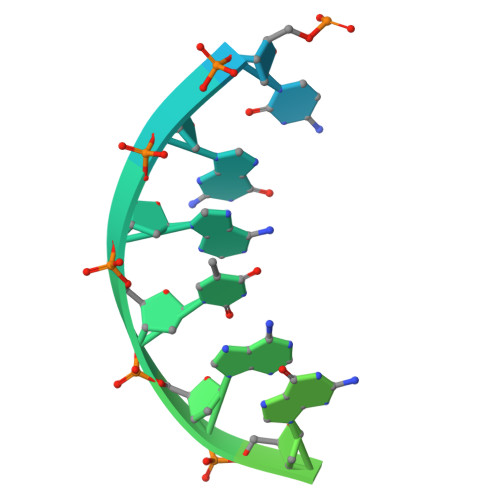
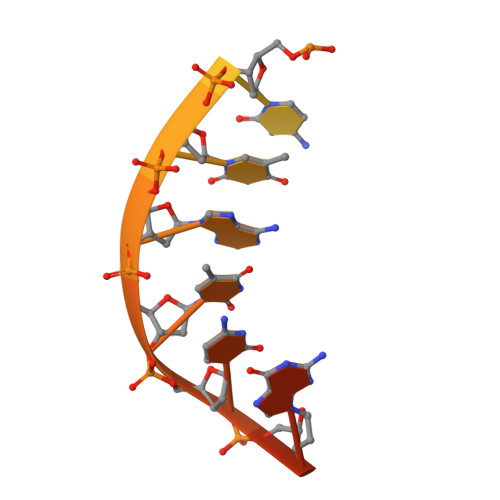
The ATP-dependent DNA ligase Lig E is present as an accessory DNA ligase in numerous proteobacterial genomes, including many disease-causing species. Here we have constructed a genomic Lig E knock-out in the obligate human pathogen Neisseria gonorrhoeae and characterised its growth and infection phenotype. This demonstrates that N. gonorrhoeae Lig E is a non-essential gene and its deletion does not cause defects in replication or survival of DNA-damaging stressors. Knock-out strains were partially defective in biofilm formation on an artificial surface as well as adhesion to epithelial cells. In addition to in vivo characterisation, we have recombinantly expressed and assayed N. gonorrhoeae Lig E and determined the crystal structure of the enzyme-adenylate engaged with DNA substrate in an open non-catalytic conformation. These findings, coupled with the predicted extracellular/ periplasmic location of Lig E indicates a role in extracellular DNA joining as well as providing insight into the binding dynamics of these minimal DNA ligases.
Organizational Affiliation:
School of Science, University of Waikato, Hamilton, New Zealand.