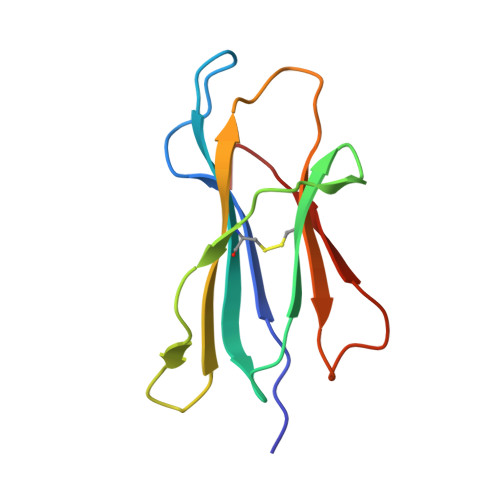
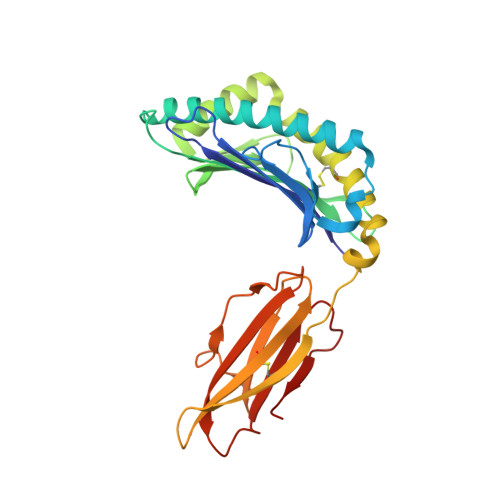
CD8 + T-cell responses towards conserved influenza B virus epitopes across anatomical sites and age.
Menon, T., Illing, P.T., Chaurasia, P., McQuilten, H.A., Shepherd, C., Rowntree, L.C., Petersen, J., Littler, D.R., Khuu, G., Huang, Z., Allen, L.F., Rockman, S., Crowe, J., Flanagan, K.L., Wakim, L.M., Nguyen, T.H.O., Mifsud, N.A., Rossjohn, J., Purcell, A.W., van de Sandt, C.E., Kedzierska, K.(2024) Nat Commun 15: 3387-3387
- PubMed: 38684663
- DOI: https://doi.org/10.1038/s41467-024-47576-y
- Primary Citation of Related Structures:
8TUB, 8TUH - PubMed Abstract:
Influenza B viruses (IBVs) cause substantive morbidity and mortality, and yet immunity towards IBVs remains understudied. CD8 + T-cells provide broadly cross-reactive immunity and alleviate disease severity by recognizing conserved epitopes. Despite the IBV burden, only 18 IBV-specific T-cell epitopes restricted by 5 HLAs have been identified currently. A broader array of conserved IBV T-cell epitopes is needed to develop effective cross-reactive T-cell based IBV vaccines. Here we identify 9 highly conserved IBV CD8 + T-cell epitopes restricted to HLA-B*07:02, HLA-B*08:01 and HLA-B*35:01. Memory IBV-specific tetramer + CD8 + T-cells are present within blood and tissues. Frequencies of IBV-specific CD8 + T-cells decline with age, but maintain a central memory phenotype. HLA-B*07:02 and HLA-B*08:01-restricted NP 30-38 epitope-specific T-cells have distinct T-cell receptor repertoires. We provide structural basis for the IBV HLA-B*07:02-restricted NS1 196-206 (11-mer) and HLA-B*07:02-restricted NP 30-38 epitope presentation. Our study increases the number of IBV CD8 + T-cell epitopes, and defines IBV-specific CD8 + T-cells at cellular and molecular levels, across tissues and age.
- Department of Microbiology and Immunology, University of Melbourne, at the Peter Doherty Institute for Infection and Immunity, Parkville, VIC, Australia.
Organizational Affiliation: