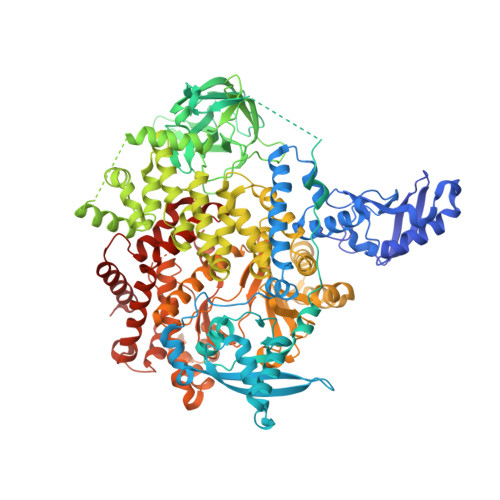
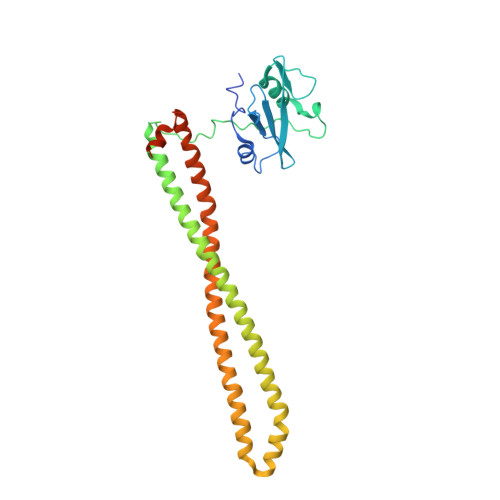
Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Varkaris, A., Pazolli, E., Gunaydin, H., Wang, Q., Pierce, L., Boezio, A.A., Bulku, A., DiPietro, L., Fridrich, C., Frost, A., Giordanetto, F., Hamilton, E.P., Harris, K., Holliday, M., Hunter, T.L., Iskandar, A., Ji, Y., Larivee, A., LaRochelle, J.R., Lescarbeau, A., Llambi, F., Lormil, B., Mader, M.M., Mar, B.G., Martin, I., McLean, T.H., Michelsen, K., Pechersky, Y., Puente-Poushnejad, E., Raynor, K., Rogala, D., Samadani, R., Schram, A.M., Shortsleeves, K., Swaminathan, S., Tajmir, S., Tan, G., Tang, Y., Valverde, R., Wehrenberg, B., Wilbur, J., Williams, B.R., Zeng, H., Zhang, H., Walters, W.P., Wolf, B.B., Shaw, D.E., Bergstrom, D.A., Watters, J., Fraser, J.S., Fortin, P.D., Kipp, D.R.(2024) Cancer Discov 14: 240-257
- PubMed: 37916956
- DOI: https://doi.org/10.1158/2159-8290.CD-23-0944
- Primary Citation of Related Structures:
8TS7, 8TS8, 8TS9, 8TSA, 8TSB, 8TSC, 8TSD, 8TU6 - PubMed Abstract:
PIK3CA (PI3Kα) is a lipid kinase commonly mutated in cancer, including ∼40% of hormone receptor-positive breast cancer. The most frequently observed mutants occur in the kinase and helical domains. Orthosteric PI3Kα inhibitors suffer from poor selectivity leading to undesirable side effects, most prominently hyperglycemia due to inhibition of wild-type (WT) PI3Kα. Here, we used molecular dynamics simulations and cryo-electron microscopy to identify an allosteric network that provides an explanation for how mutations favor PI3Kα activation. A DNA-encoded library screen leveraging electron microscopy-optimized constructs, differential enrichment, and an orthosteric-blocking compound led to the identification of RLY-2608, a first-in-class allosteric mutant-selective inhibitor of PI3Kα. RLY-2608 inhibited tumor growth in PIK3CA-mutant xenograft models with minimal impact on insulin, a marker of dysregulated glucose homeostasis. RLY-2608 elicited objective tumor responses in two patients diagnosed with advanced hormone receptor-positive breast cancer with kinase or helical domain PIK3CA mutations, with no observed WT PI3Kα-related toxicities. Treatments for PIK3CA-mutant cancers are limited by toxicities associated with the inhibition of WT PI3Kα. Molecular dynamics, cryo-electron microscopy, and DNA-encoded libraries were used to develop RLY-2608, a first-in-class inhibitor that demonstrates mutant selectivity in patients. This marks the advance of clinical mutant-selective inhibition that overcomes limitations of orthosteric PI3Kα inhibitors. See related commentary by Gong and Vanhaesebroeck, p. 204 . See related article by Varkaris et al., p. 227 . This article is featured in Selected Articles from This Issue, p. 201.
- Mass General Cancer Center and Department of Medicine, Harvard Medical School, Boston, Massachusetts.
Organizational Affiliation: