Structural basis of human 20S proteasome biogenesis.
Zhang, H., Zhou, C., Mohammad, Z., Zhao, J.(2024) Nat Commun 15: 8184-8184
- PubMed: 39294158
- DOI: https://doi.org/10.1038/s41467-024-52513-0
- Primary Citation of Related Structures:
8TM3, 8TM4, 8TM5, 8TM6 - PubMed Abstract:
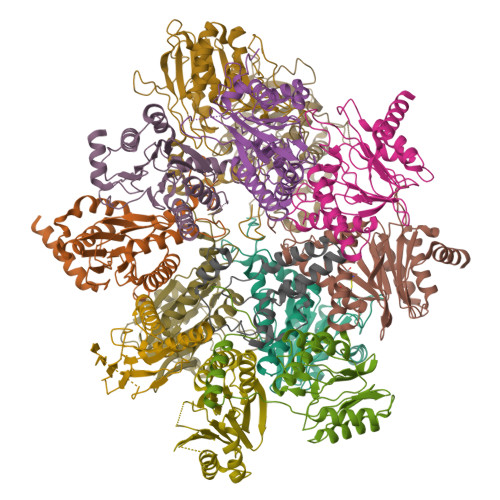
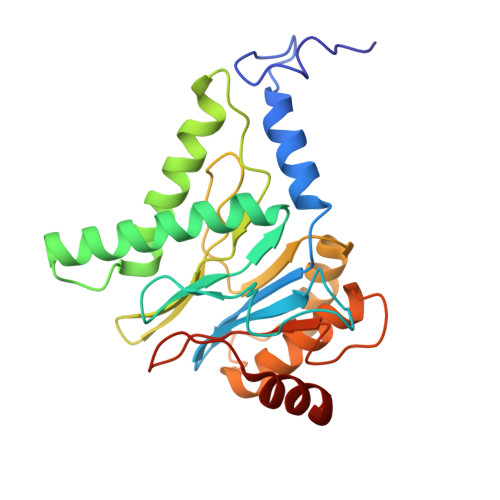
New proteasomes are produced to accommodate increases in cellular catabolic demand and prevent the accumulation of cytotoxic proteins. Formation of the proteasomal 20S core complex relies on the function of the five chaperones PAC1-4 and POMP. Here, to understand how these chaperones facilitate proteasome assembly, we tagged the endogenous chaperones using CRISPR/Cas gene editing and examined the chaperone-bound complexes by cryo-EM. We observe an early α-ring intermediate subcomplex that is stabilized by PAC1-4, which transitions to β-ring assembly upon dissociation of PAC3/PAC4 and rearrangement of the PAC1 N-terminal tail. Completion of the β-ring and dimerization of half-proteasomes repositions critical lysine K33 to trigger cleavage of the β pro-peptides, leading to the concerted dissociation of POMP and PAC1/PAC2 to yield mature 20S proteasomes. This study reveals structural insights into critical points along the assembly pathway of the human proteasome and provides a molecular blueprint for 20S biogenesis.
Organizational Affiliation:
Cancer Metabolism and Microenvironment Program, Sanford Burnham Prebys Medical Discovery Institute, La Jolla, 92037, USA.