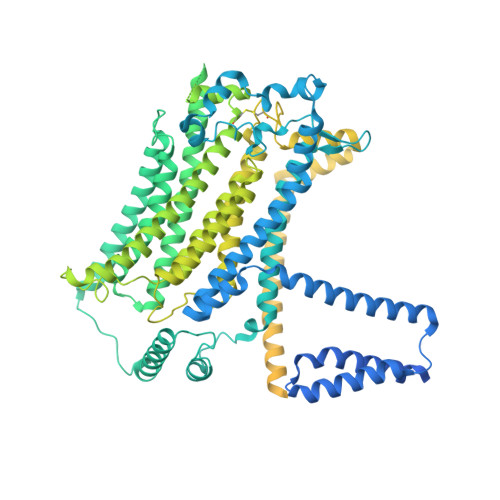
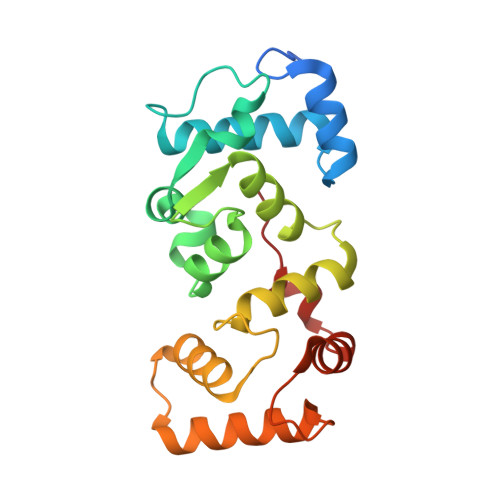
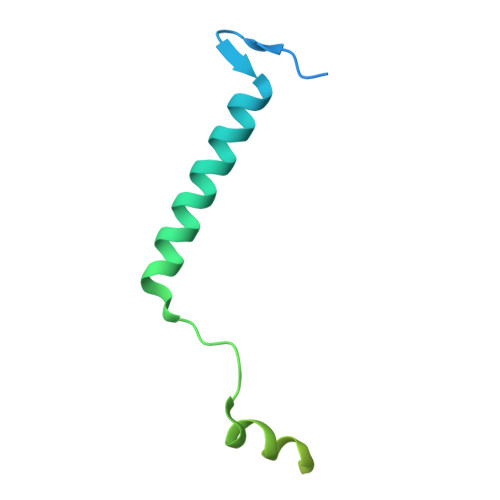
The structure of the Caenorhabditis elegans TMC-2 complex suggests roles of lipid-mediated subunit contacts in mechanosensory transduction.
Clark, S., Jeong, H., Posert, R., Goehring, A., Gouaux, E.(2024) Proc Natl Acad Sci U S A 121: e2314096121-e2314096121
- PubMed: 38354260
- DOI: https://doi.org/10.1073/pnas.2314096121
- Primary Citation of Related Structures:
8TKP - PubMed Abstract:
Mechanotransduction is the process by which a mechanical force, such as touch, is converted into an electrical signal. Transmembrane channel-like (TMC) proteins are an evolutionarily conserved family of membrane proteins whose function has been linked to a variety of mechanosensory processes, including hearing and balance sensation in vertebrates and locomotion in Drosophila . TMC1 and TMC2 are components of ion channel complexes, but the molecular features that tune these complexes to diverse mechanical stimuli are unknown. Caenorhabditis elegans express two TMC homologs, TMC-1 and TMC-2, both of which are the likely pore-forming subunits of mechanosensitive ion channels but differ in their expression pattern and functional role in the worm. Here, we present the single-particle cryo-electron microscopy structure of the native TMC-2 complex isolated from C. elegans . The complex is composed of two copies of the pore-forming TMC-2 subunit, the calcium and integrin binding protein CALM-1 and the transmembrane inner ear protein TMIE. Comparison of the TMC-2 complex to the recently published cryo-EM structure of the C. elegans TMC-1 complex highlights conserved protein-lipid interactions, as well as a π-helical structural motif in the pore-forming helices, that together suggest a mechanism for TMC-mediated mechanosensory transduction.
- Vollum Institute, Oregon Health and Science University, Portland, OR 97239.
Organizational Affiliation: