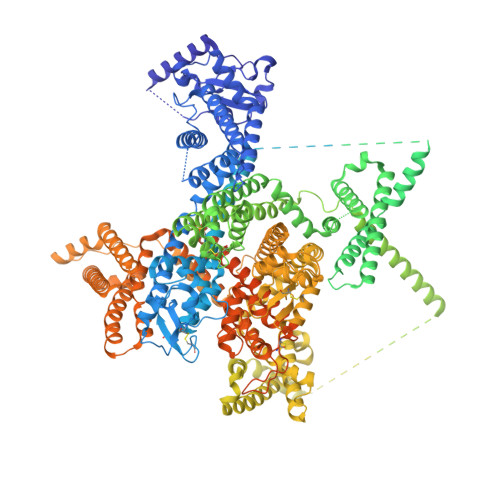
Dual-pocket inhibition of Na v channels by the antiepileptic drug lamotrigine.
Huang, J., Fan, X., Jin, X., Teng, L., Yan, N.(2023) Proc Natl Acad Sci U S A 120: e2309773120-e2309773120
- PubMed: 37782796
- DOI: https://doi.org/10.1073/pnas.2309773120
- Primary Citation of Related Structures:
8THG, 8THH - PubMed Abstract:
Voltage-gated sodium (Na v ) channels govern membrane excitability, thus setting the foundation for various physiological and neuronal processes. Na v channels serve as the primary targets for several classes of widely used and investigational drugs, including local anesthetics, antiepileptic drugs, antiarrhythmics, and analgesics. In this study, we present cryogenic electron microscopy (cryo-EM) structures of human Na v 1.7 bound to two clinical drugs, riluzole (RLZ) and lamotrigine (LTG), at resolutions of 2.9 Å and 2.7 Å, respectively. A 3D EM reconstruction of ligand-free Na v 1.7 was also obtained at 2.1 Å resolution. RLZ resides in the central cavity of the pore domain and is coordinated by residues from repeats III and IV. Whereas one LTG molecule also binds to the central cavity, the other is found beneath the intracellular gate, known as site BIG. Therefore, LTG, similar to lacosamide and cannabidiol, blocks Na v channels via a dual-pocket mechanism. These structures, complemented with docking and mutational analyses, also explain the structure-activity relationships of the LTG-related linear 6,6 series that have been developed for improved efficacy and subtype specificity on different Na v channels. Our findings reveal the molecular basis for these drugs' mechanism of action and will aid the development of novel antiepileptic and pain-relieving drugs.
- Department of Molecular Biology, Princeton University, Princeton, NJ 08544.
Organizational Affiliation: