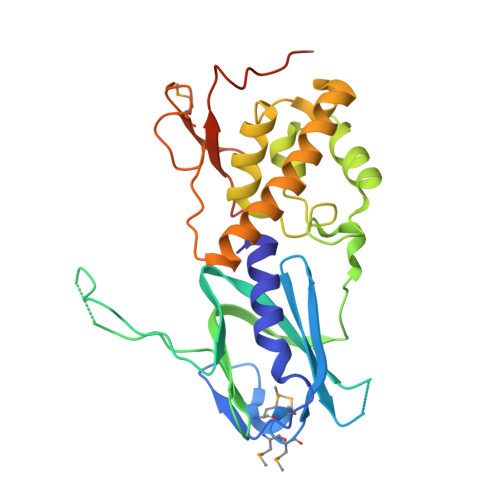
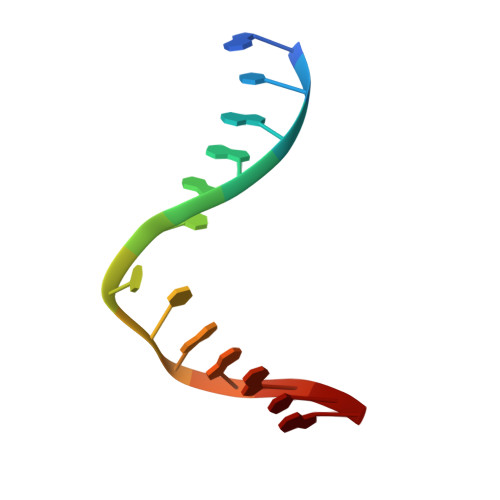
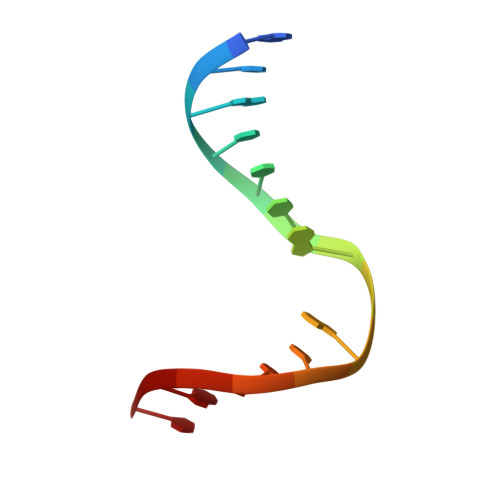
Structural and biochemical insights into NEIL2's preference for abasic sites.
Eckenroth, B.E., Bumgarner, J.D., Matsumoto-Elliott, O., David, S.S., Doublie, S.(2023) Nucleic Acids Res 51: 12508-12521
- PubMed: 37971311
- DOI: https://doi.org/10.1093/nar/gkad1075
- Primary Citation of Related Structures:
8TH9 - PubMed Abstract:
Cellular DNA is subject to damage from a multitude of sources and repair or bypass of sites of damage utilize an array of context or cell cycle dependent systems. The recognition and removal of oxidatively damaged bases is the task of DNA glycosylases from the base excision repair pathway utilizing two structural families that excise base lesions in a wide range of DNA contexts including duplex, single-stranded and bubble structures arising during transcription. The mammalian NEIL2 glycosylase of the Fpg/Nei family excises lesions from each of these DNA contexts favoring the latter two with a preference for oxidized cytosine products and abasic sites. We have determined the first liganded crystal structure of mammalian NEIL2 in complex with an abasic site analog containing DNA duplex at 2.08 Å resolution. Comparison to the unliganded structure revealed a large interdomain conformational shift upon binding the DNA substrate accompanied by local conformational changes in the C-terminal domain zinc finger and N-terminal domain void-filling loop necessary to position the enzyme on the DNA. The detailed biochemical analysis of NEIL2 with an array of oxidized base lesions indicates a significant preference for its lyase activity likely to be paramount when interpreting the biological consequences of variants.
- Department of Microbiology and Molecular Genetics, University of Vermont, Stafford Hall, 95 Carrigan Drive, Burlington, VT 05405, USA.
Organizational Affiliation: