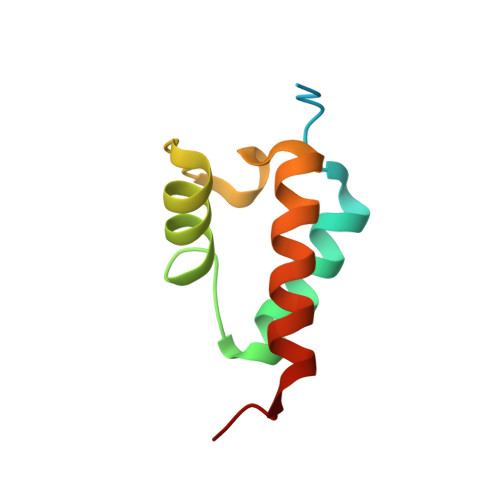
Structural Basis for the Interaction of Red beta Single-Strand Annealing Protein with Escherichia coli Single-Stranded DNA-Binding Protein.
Zakharova, K., Liu, M., Greenwald, J.R., Caldwell, B.C., Qi, Z., Wysocki, V.H., Bell, C.E.(2024) J Mol Biology 436: 168590-168590
- PubMed: 38663547
- DOI: https://doi.org/10.1016/j.jmb.2024.168590
- Primary Citation of Related Structures:
8TFU, 8TG7, 8TG8, 8TGC - PubMed Abstract:
Redβ is a protein from bacteriophage λ that binds to single-stranded DNA (ssDNA) to promote the annealing of complementary strands. Together with λ-exonuclease (λ-exo), Redβ is part of a two-component DNA recombination system involved in multiple aspects of genome maintenance. The proteins have been exploited in powerful methods for bacterial genome engineering in which Redβ can anneal an electroporated oligonucleotide to a complementary target site at the lagging strand of a replication fork. Successful annealing in vivo requires the interaction of Redβ with E. coli single-stranded DNA-binding protein (SSB), which coats the ssDNA at the lagging strand to coordinate access of numerous replication proteins. Previous mutational analysis revealed that the interaction between Redβ and SSB involves the C-terminal domain (CTD) of Redβ and the C-terminal tail of SSB (SSB-Ct), the site for binding of numerous host proteins. Here, we have determined the x-ray crystal structure of Redβ CTD in complex with a peptide corresponding to the last nine residues of SSB (MDFDDDIPF). Formation of the complex is predominantly mediated by hydrophobic interactions between two phenylalanine side chains of SSB (Phe-171 and Phe-177) and an apolar groove on the CTD, combined with electrostatic interactions between the C-terminal carboxylate of SSB and Lys-214 of the CTD. Mutation of any of these residues to alanine significantly disrupts the interaction of full-length Redβ and SSB proteins. Structural knowledge of this interaction will help to expand the utility of Redβ-mediated recombination to a wider range of bacterial hosts for applications in synthetic biology.
- Department of Biological Chemistry and Pharmacology, The Ohio State University, Columbus, OH, USA.
Organizational Affiliation: