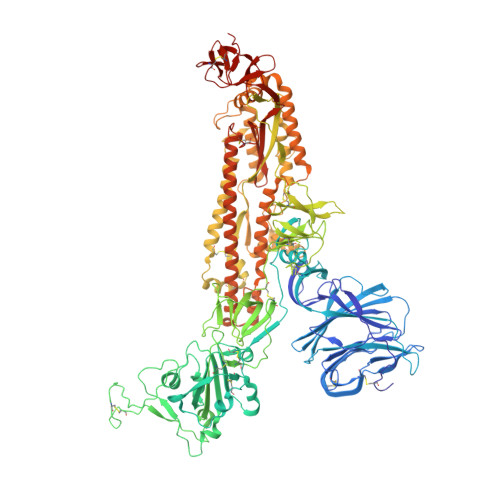
Variation in structural motifs within SARS-related coronavirus spike proteins.
Hills, F.R., Eruera, A.R., Hodgkinson-Bean, J., Jorge, F., Easingwood, R., Brown, S.H.J., Bouwer, J.C., Li, Y.P., Burga, L.N., Bostina, M.(2024) PLoS Pathog 20: e1012158-e1012158
- PubMed: 38805567
- DOI: https://doi.org/10.1371/journal.ppat.1012158
- Primary Citation of Related Structures:
8TC0, 8TC1, 8TC5 - PubMed Abstract:
SARS-CoV-2 is the third known coronavirus (CoV) that has crossed the animal-human barrier in the last two decades. However, little structural information exists related to the close genetic species within the SARS-related coronaviruses. Here, we present three novel SARS-related CoV spike protein structures solved by single particle cryo-electron microscopy analysis derived from bat (bat SL-CoV WIV1) and civet (cCoV-SZ3, cCoV-007) hosts. We report complex glycan trees that decorate the glycoproteins and density for water molecules which facilitated modeling of the water molecule coordination networks within structurally important regions. We note structural conservation of the fatty acid binding pocket and presence of a linoleic acid molecule which are associated with stabilization of the receptor binding domains in the "down" conformation. Additionally, the N-terminal biliverdin binding pocket is occupied by a density in all the structures. Finally, we analyzed structural differences in a loop of the receptor binding motif between coronaviruses known to infect humans and the animal coronaviruses described in this study, which regulate binding to the human angiotensin converting enzyme 2 receptor. This study offers a structural framework to evaluate the close relatives of SARS-CoV-2, the ability to inform pandemic prevention, and aid in the development of pan-neutralizing treatments.
- Department of Microbiology and Immunology, University of Otago, Dunedin, New Zealand.
Organizational Affiliation: