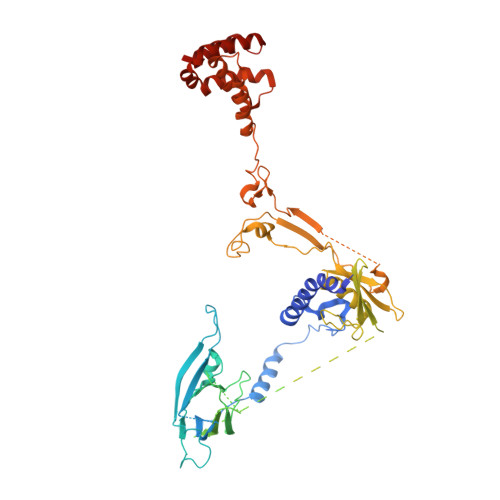
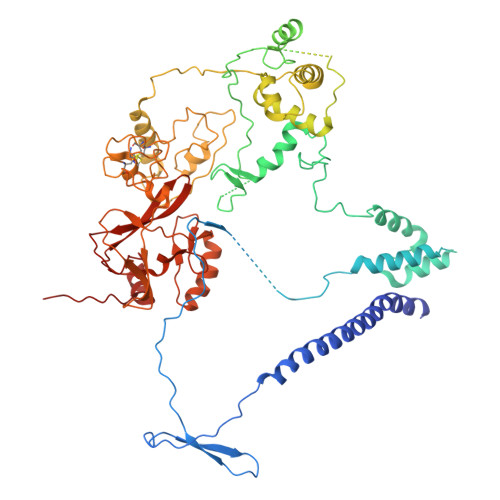
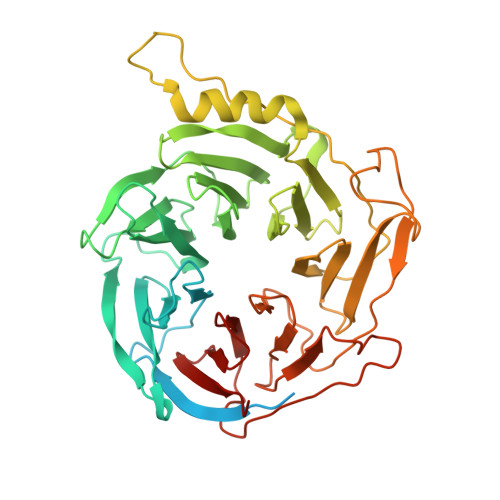
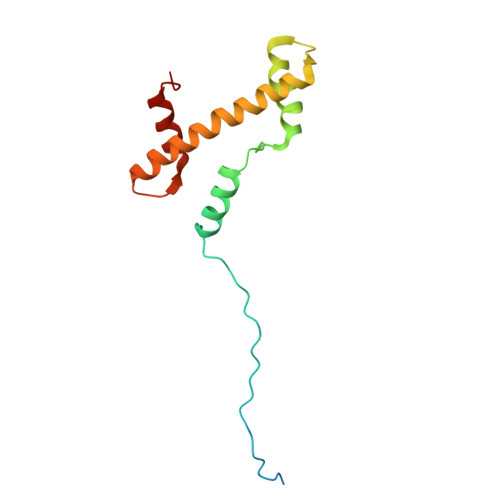
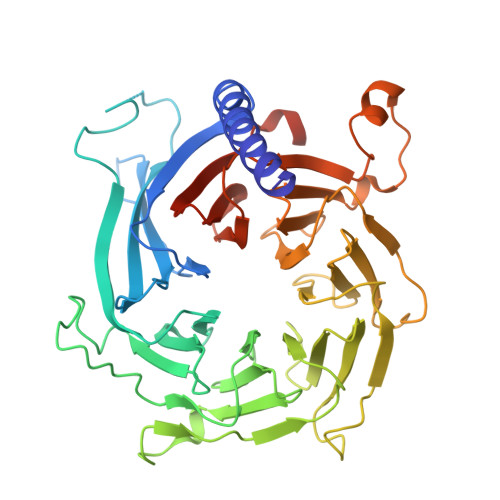
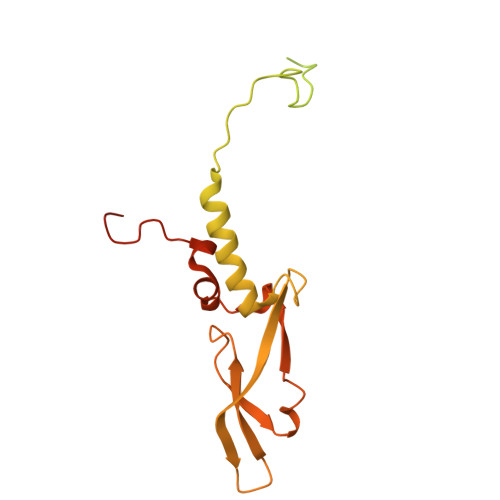
Activation of automethylated PRC2 by dimerization on chromatin.
Sauer, P.V., Pavlenko, E., Cookis, T., Zirden, L.C., Renn, J., Singhal, A., Hunold, P., Hoehne-Wiechmann, M.N., van Ray, O., Kaschani, F., Kaiser, M., Hansel-Hertsch, R., Sanbonmatsu, K.Y., Nogales, E., Poepsel, S.(2024) Mol Cell 84: 3885-3898.e8
- PubMed: 39303719
- DOI: https://doi.org/10.1016/j.molcel.2024.08.025
- Primary Citation of Related Structures:
8T9G, 8TAS, 8TB9 - PubMed Abstract:
Polycomb repressive complex 2 (PRC2) is an epigenetic regulator that trimethylates lysine 27 of histone 3 (H3K27me3) and is essential for embryonic development and cellular differentiation. H3K27me3 is associated with transcriptionally repressed chromatin and is established when PRC2 is allosterically activated upon methyl-lysine binding by the regulatory subunit EED. Automethylation of the catalytic subunit enhancer of zeste homolog 2 (EZH2) stimulates its activity by an unknown mechanism. Here, we show that human PRC2 forms a dimer on chromatin in which an inactive, automethylated PRC2 protomer is the allosteric activator of a second PRC2 that is poised to methylate H3 of a substrate nucleosome. Functional assays support our model of allosteric trans-autoactivation via EED, suggesting a previously unknown mechanism mediating context-dependent activation of PRC2. Our work showcases the molecular mechanism of auto-modification-coupled dimerization in the regulation of chromatin-modifying complexes.
- California Institute for Quantitative Biology (QB3), University of California, Berkeley, CA 94720, USA; Howard Hughes Medical Institute, University of California, Berkeley, CA 94720, USA.
Organizational Affiliation: