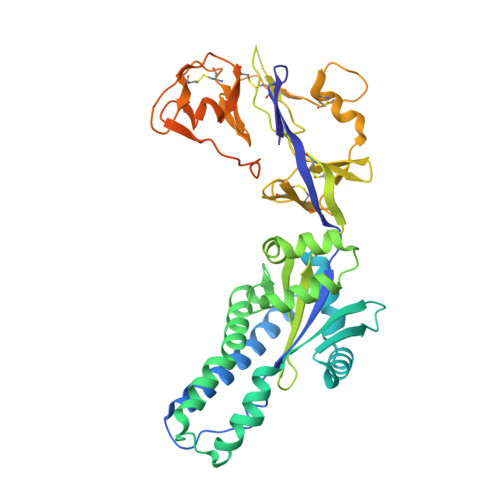
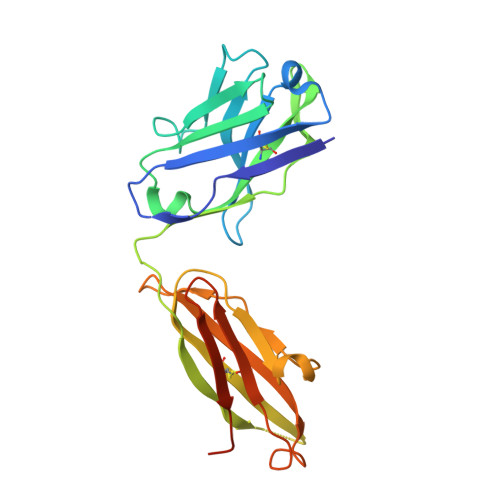
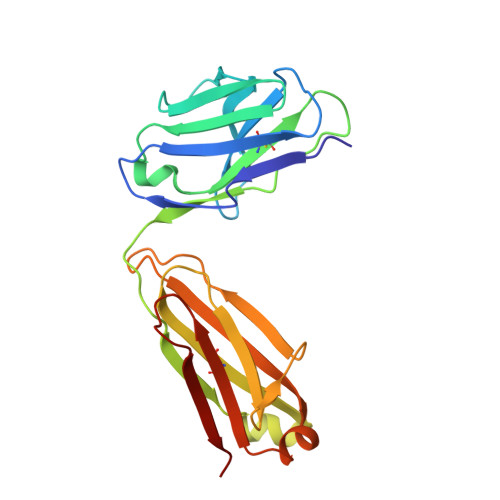
Structural characterization of M8C10, a neutralizing antibody targeting a highly conserved prefusion-specific epitope on the metapneumovirus fusion trimerization interface.
Xiao, X., Wen, Z., Chen, Q., Shipman, J.M., Kostas, J., Reid, J.C., Warren, C., Tang, A., Luo, B., O'Donnell, G., Fridman, A., Chen, Z., Vora, K.A., Zhang, L., Su, H.-P., Eddins, M.J.(2023) J Virol 97: e0105223-e0105223
- PubMed: 38032197
- DOI: https://doi.org/10.1128/jvi.01052-23
- Primary Citation of Related Structures:
8T9Z - PubMed Abstract:
Human metapneumovirus (hMPV) is a common pathogen causing lower respiratory tract infections worldwide and can develop severe symptoms in high-risk populations such as infants, the elderly, and immunocompromised patients. There are no approved hMPV vaccines or neutralizing antibodies available for therapeutic or prophylactic use. The trimeric hMPV fusion F protein is the major target of neutralizing antibodies in human sera. Understanding the immune recognition of antibodies to hMPV-F antigen will provide critical insights into developing efficacious hMPV monoclonal antibodies and vaccines.
- Infectious Diseases and Vaccines Discovery, Merck & Co., Inc., West Point, Pennsylvania, USA.
Organizational Affiliation: