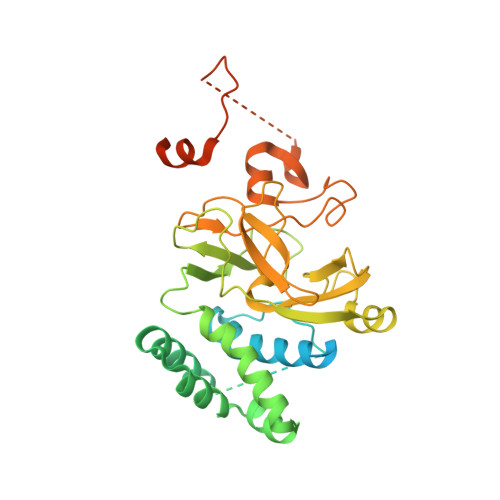
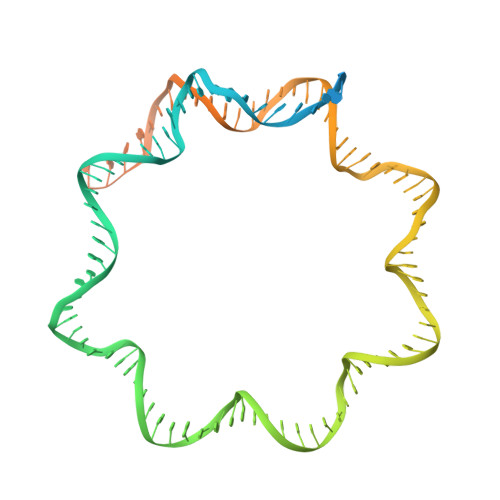
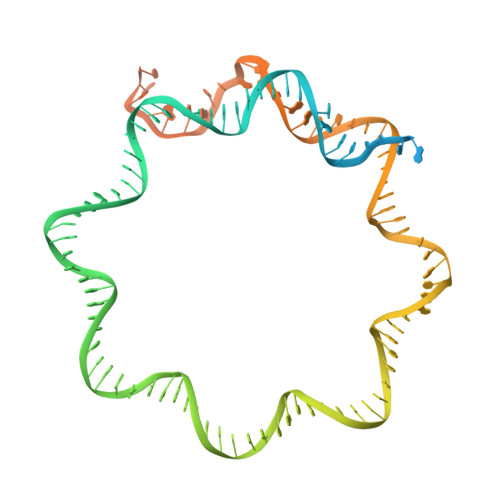
Catalytic and non-catalytic mechanisms of histone H4 lysine 20 methyltransferase SUV420H1.
Abini-Agbomson, S., Gretarsson, K., Shih, R.M., Hsieh, L., Lou, T., De Ioannes, P., Vasilyev, N., Lee, R., Wang, M., Simon, M.D., Armache, J.P., Nudler, E., Narlikar, G., Liu, S., Lu, C., Armache, K.J.(2023) Mol Cell 83: 2872-2883.e7
- PubMed: 37595555
- DOI: https://doi.org/10.1016/j.molcel.2023.07.020
- Primary Citation of Related Structures:
8T9F, 8T9H, 8THU - PubMed Abstract:
SUV420H1 di- and tri-methylates histone H4 lysine 20 (H4K20me2/H4K20me3) and plays crucial roles in DNA replication, repair, and heterochromatin formation. It is dysregulated in several cancers. Many of these processes were linked to its catalytic activity. However, deletion and inhibition of SUV420H1 have shown distinct phenotypes, suggesting that the enzyme likely has uncharacterized non-catalytic activities. Our cryoelectron microscopy (cryo-EM), biochemical, biophysical, and cellular analyses reveal how SUV420H1 recognizes its nucleosome substrates, and how histone variant H2A.Z stimulates its catalytic activity. SUV420H1 binding to nucleosomes causes a dramatic detachment of nucleosomal DNA from the histone octamer, which is a non-catalytic activity. We hypothesize that this regulates the accessibility of large macromolecular complexes to chromatin. We show that SUV420H1 can promote chromatin condensation, another non-catalytic activity that we speculate is needed for its heterochromatin functions. Together, our studies uncover and characterize the catalytic and non-catalytic mechanisms of SUV420H1, a key histone methyltransferase that plays an essential role in genomic stability.
- Department of Biochemistry and Molecular Pharmacology, New York University Grossman School of Medicine, New York, NY, USA.
Organizational Affiliation: