Rapid affinity optimization of an anti-TREM2 clinical lead antibody by cross-lineage immune repertoire mining.
Hsiao, Y.C., Wallweber, H.A., Alberstein, R.G., Lin, Z., Du, C., Etxeberria, A., Aung, T., Shang, Y., Seshasayee, D., Seeger, F., Watkins, A.M., Hansen, D.V., Bohlen, C.J., Hsu, P.L., Hotzel, I.(2024) Nat Commun 15: 8382-8382
- PubMed: 39333507
- DOI: https://doi.org/10.1038/s41467-024-52442-y
- Primary Citation of Related Structures:
8T51, 8T59 - PubMed Abstract:
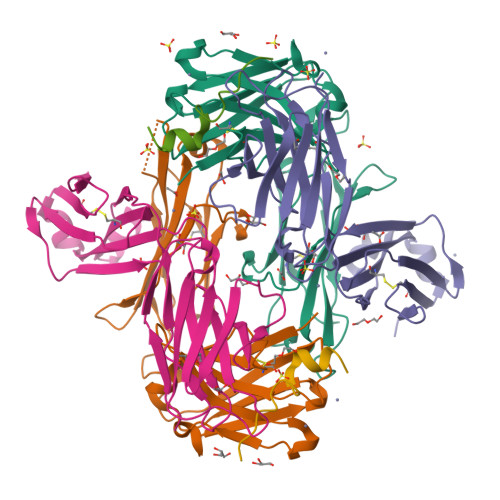
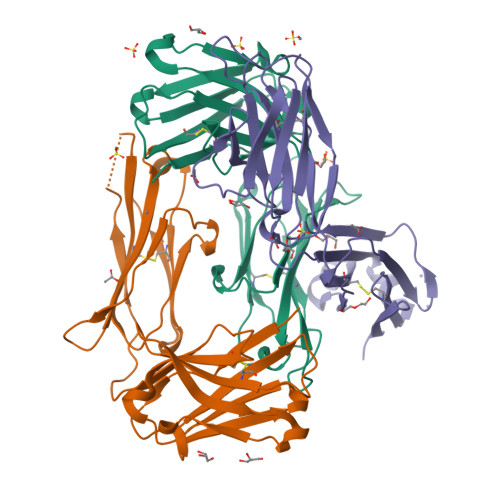
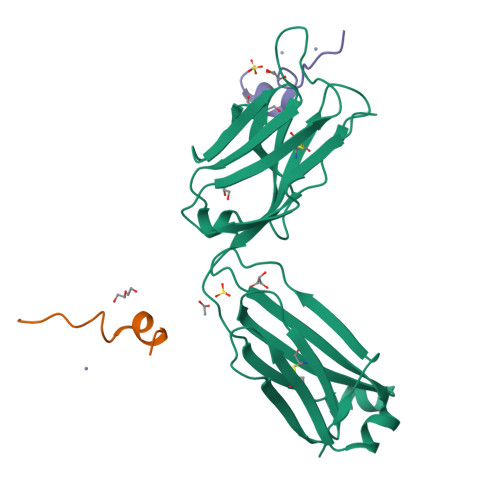
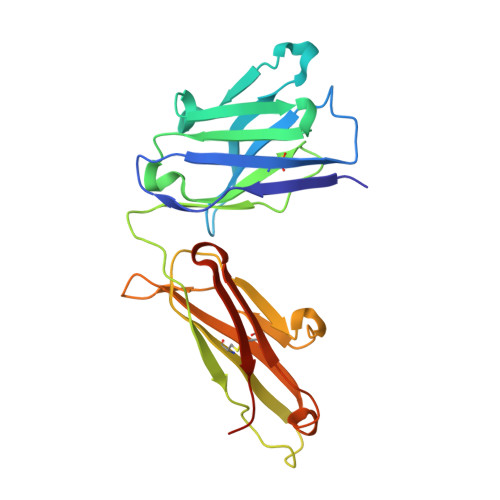
We describe a process for rapid antibody affinity optimization by repertoire mining to identify clones across B cell clonal lineages based on convergent immune responses where antigen-specific clones with the same heavy (V H ) and light chain germline segment pairs, or parallel lineages, bind a single epitope on the antigen. We use this convergence framework to mine unique and distinct V H lineages from rat anti-triggering receptor on myeloid cells 2 (TREM2) antibody repertoire datasets with high diversity in the third complementarity-determining loop region (CDR H3) to further affinity-optimize a high-affinity agonistic anti-TREM2 antibody while retaining critical functional properties. Structural analyses confirm a nearly identical binding mode of anti-TREM2 variants with subtle but significant structural differences in the binding interface. Parallel lineage repertoire mining is uniquely tailored to rationally explore the large CDR H3 sequence space in antibody repertoires and can be easily and generally applied to antibodies discovered in vivo.
Organizational Affiliation:
Department of Antibody Engineering, Genentech, South San Francisco, CA, 94080, USA.