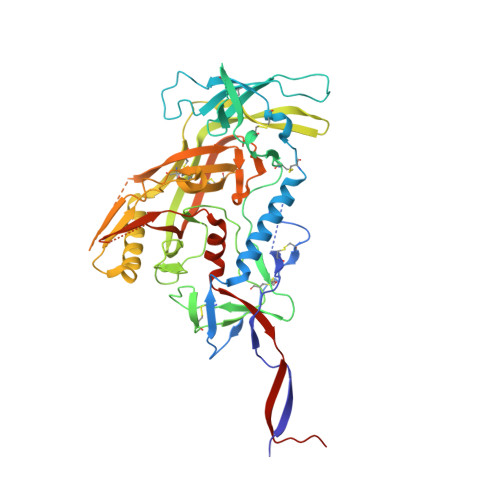
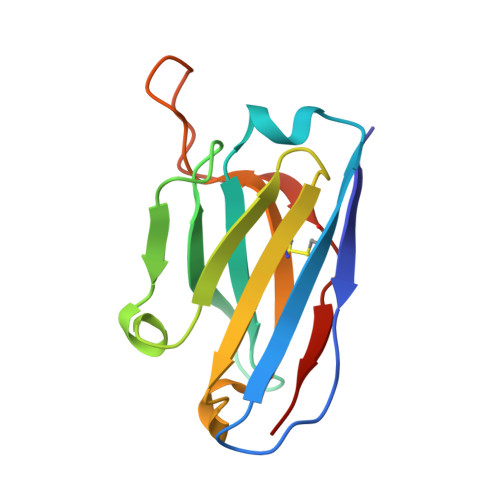
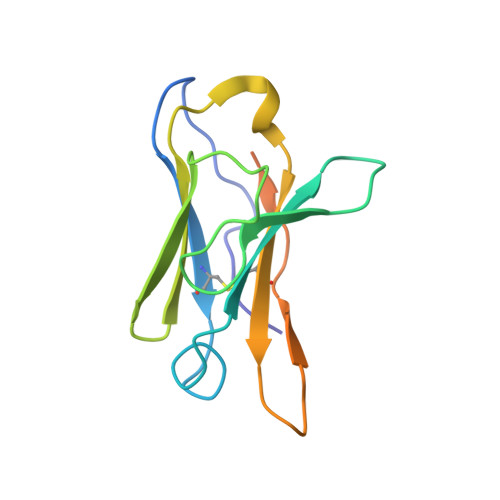
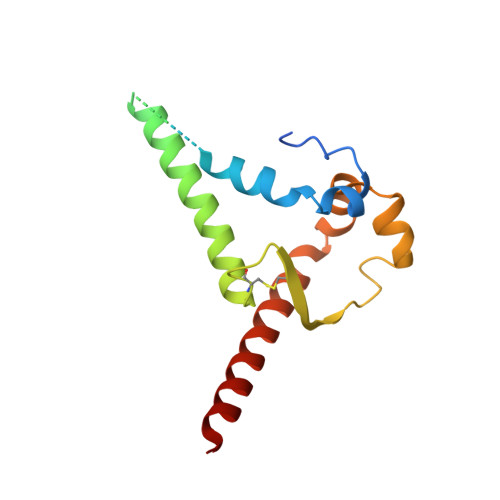
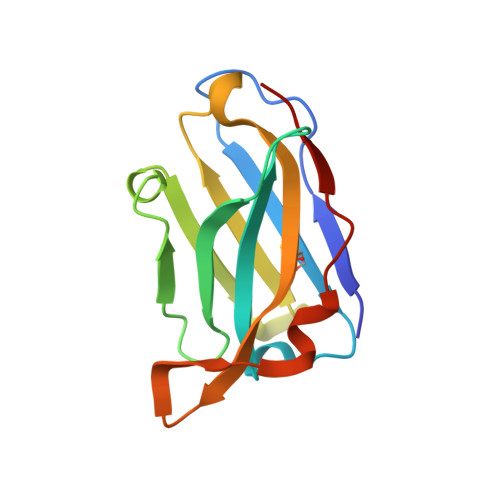
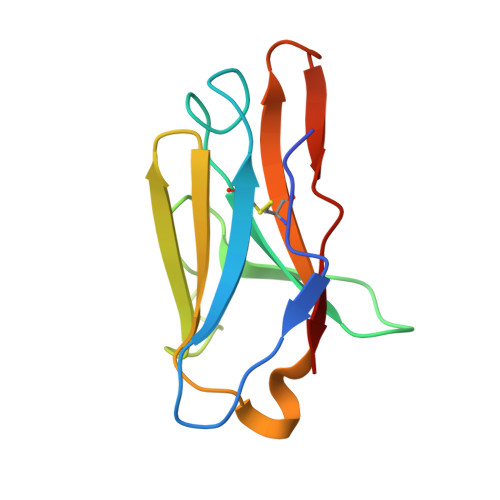
Vaccine priming of rare HIV broadly neutralizing antibody precursors in nonhuman primates.
Steichen, J.M., Phung, I., Salcedo, E., Ozorowski, G., Willis, J.R., Baboo, S., Liguori, A., Cottrell, C.A., Torres, J.L., Madden, P.J., Ma, K.M., Sutton, H.J., Lee, J.H., Kalyuzhniy, O., Allen, J.D., Rodriguez, O.L., Adachi, Y., Mullen, T.M., Georgeson, E., Kubitz, M., Burns, A., Barman, S., Mopuri, R., Metz, A., Altheide, T.K., Diedrich, J.K., Saha, S., Shields, K., Schultze, S.E., Smith, M.L., Schiffner, T., Burton, D.R., Watson, C.T., Bosinger, S.E., Crispin, M., Yates 3rd, J.R., Paulson, J.C., Ward, A.B., Sok, D., Crotty, S., Schief, W.R.(2024) Science 384: eadj8321-eadj8321
- PubMed: 38753769
- DOI: https://doi.org/10.1126/science.adj8321
- Primary Citation of Related Structures:
8T49, 8T4A, 8T4B, 8T4D, 8T4K, 8T4L - PubMed Abstract:
Germline-targeting immunogens hold promise for initiating the induction of broadly neutralizing antibodies (bnAbs) to HIV and other pathogens. However, antibody-antigen recognition is typically dominated by heavy chain complementarity determining region 3 (HCDR3) interactions, and vaccine priming of HCDR3-dominant bnAbs by germline-targeting immunogens has not been demonstrated in humans or outbred animals. In this work, immunization with N332-GT5, an HIV envelope trimer designed to target precursors of the HCDR3-dominant bnAb BG18, primed bnAb-precursor B cells in eight of eight rhesus macaques to substantial frequencies and with diverse lineages in germinal center and memory B cells. We confirmed bnAb-mimicking, HCDR3-dominant, trimer-binding interactions with cryo-electron microscopy. Our results demonstrate proof of principle for HCDR3-dominant bnAb-precursor priming in outbred animals and suggest that N332-GT5 holds promise for the induction of similar responses in humans.
- Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: