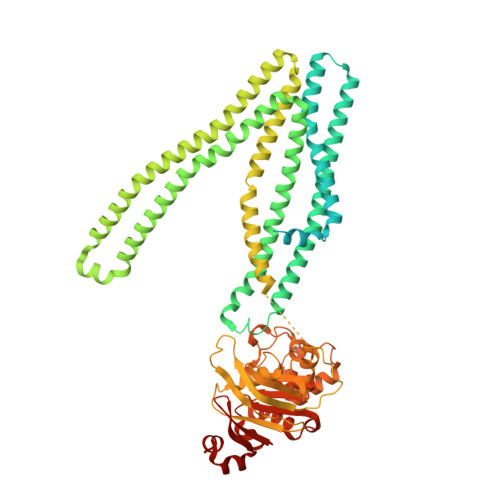
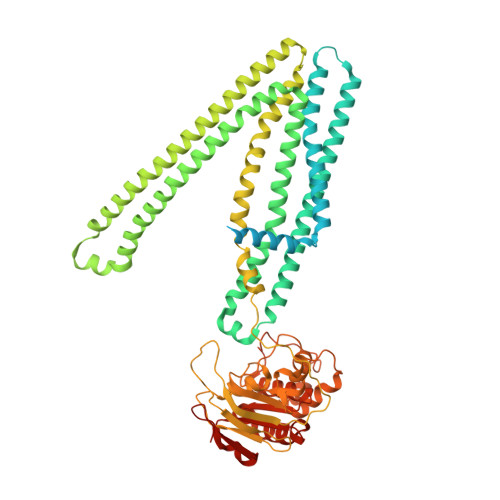
Principles of peptide selection by the transporter associated with antigen processing.
Lee, J., Oldham, M.L., Manon, V., Chen, J.(2024) Proc Natl Acad Sci U S A 121: e2320879121-e2320879121
- PubMed: 38805290
- DOI: https://doi.org/10.1073/pnas.2320879121
- Primary Citation of Related Structures:
8T46, 8T4E, 8T4F, 8T4G, 8T4H, 8T4I, 8T4J - PubMed Abstract:
Our ability to fight pathogens relies on major histocompatibility complex class I (MHC-I) molecules presenting diverse antigens on the surface of diseased cells. The transporter associated with antigen processing (TAP) transports nearly the entire repertoire of antigenic peptides into the endoplasmic reticulum for MHC-I loading. How TAP transports peptides specific for MHC-I is unclear. In this study, we used cryo-EM to determine a series of structures of human TAP, both in the absence and presence of peptides with various sequences and lengths. The structures revealed that peptides of eight or nine residues in length bind in a similarly extended conformation, despite having little sequence overlap. We also identified two peptide-anchoring pockets on either side of the transmembrane cavity, each engaging one end of a peptide with primarily main chain atoms. Occupation of both pockets results in a global conformational change in TAP, bringing the two halves of the transporter closer together to prime it for isomerization and ATP hydrolysis. Shorter peptides are able to bind to each pocket separately but are not long enough to bridge the cavity to bind to both simultaneously. Mutations that disrupt hydrogen bonds with the N and C termini of peptides almost abolish MHC-I surface expression. Our findings reveal that TAP functions as a molecular caliper that selects peptides according to length rather than sequence, providing antigen diversity for MHC-I presentation.
- Laboratory of Membrane Biophysics and Biology, The Rockefeller University, New York, NY 10065.
Organizational Affiliation: