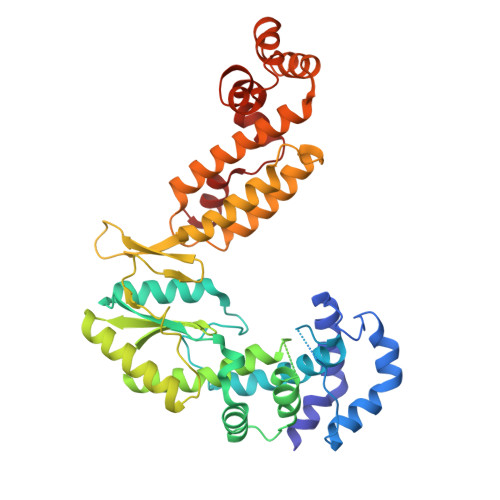
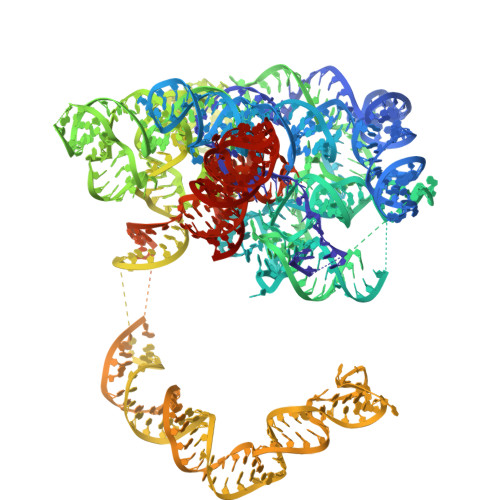
Structural insights into intron catalysis and dynamics during splicing.
Xu, L., Liu, T., Chung, K., Pyle, A.M.(2023) Nature 624: 682-688
- PubMed: 37993708
- DOI: https://doi.org/10.1038/s41586-023-06746-6
- Primary Citation of Related Structures:
8T2R, 8T2S, 8T2T - PubMed Abstract:
The group II intron ribonucleoprotein is an archetypal splicing system with numerous mechanistic parallels to the spliceosome, including excision of lariat introns 1,2 . Despite the importance of branching in RNA metabolism, structural understanding of this process has remained elusive. Here we present a comprehensive analysis of three single-particle cryogenic electron microscopy structures captured along the splicing pathway. They reveal the network of molecular interactions that specifies the branchpoint adenosine and positions key functional groups to catalyse lariat formation and coordinate exon ligation. The structures also reveal conformational rearrangements of the branch helix and the mechanism of splice site exchange that facilitate the transition from branching to ligation. These findings shed light on the evolution of splicing and highlight the conservation of structural components, catalytic mechanism and dynamical strategies retained through time in premessenger RNA splicing machines.
- Howard Hughes Medical Institute, Chevy Chase, MD, USA. ling.xu@yale.edu.
Organizational Affiliation: