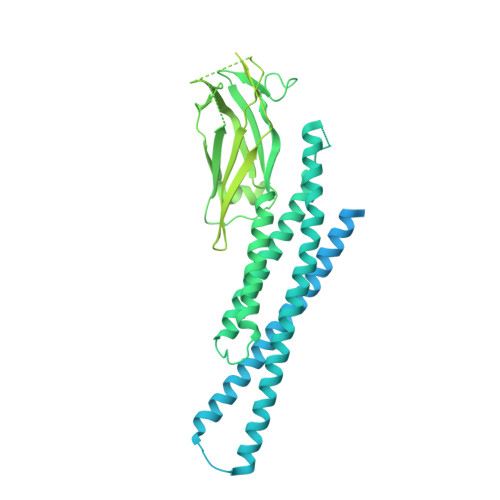
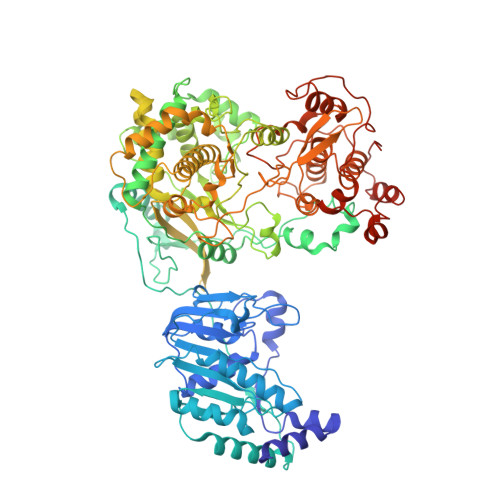
A conformational selection mechanism of flavivirus NS5 for species-specific STAT2 inhibition.
Biswal, M., Yao, W., Lu, J., Chen, J., Morrison, J., Hai, R., Song, J.(2024) Commun Biol 7: 76-76
- PubMed: 38195857
- DOI: https://doi.org/10.1038/s42003-024-05768-8
- Primary Citation of Related Structures:
8T12, 8T13 - PubMed Abstract:
Flaviviruses, including Zika virus (ZIKV) and Dengue virus (DENV), rely on their non-structural protein 5 (NS5) for both replication of viral genome and suppression of host IFN signaling. DENV and ZIKV NS5s were shown to facilitate proteosome-mediated protein degradation of human STAT2 (hSTAT2). However, how flavivirus NS5s have evolved for species-specific IFN-suppression remains unclear. Here we report structure-function characterization of the DENV serotype 2 (DENV2) NS5-hSTAT2 complex. The MTase and RdRP domains of DENV2 NS5 form an extended conformation to interact with the coiled-coil and N-terminal domains of hSTAT2, thereby promoting hSTAT2 degradation in cells. Disruption of the extended conformation of DENV2/ZIKV NS5, but not the alternative compact state, impaired their hSTAT2 binding. Our comparative structural analysis of flavivirus NS5s further reveals a conserved protein-interaction platform with subtle amino-acid variations likely underpinning diverse IFN-suppression mechanisms. Together, this study uncovers a conformational selection mechanism underlying species-specific hSTAT2 inhibition by flavivirus NS5.
- Department of Biochemistry, University of California, Riverside, CA, USA.
Organizational Affiliation: