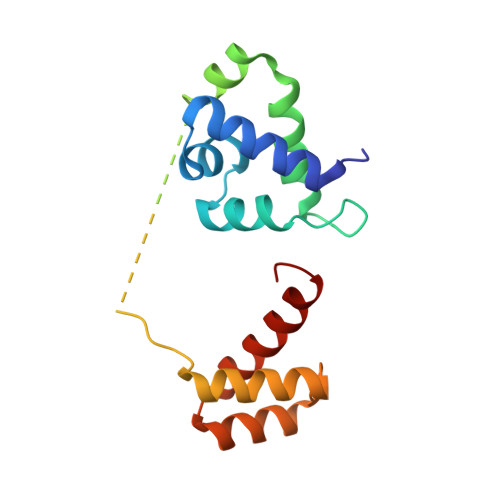
The homeodomain regulates stable DNA binding of prostate cancer target ONECUT2.
Chatterjee, A., Gallent, B., Katiki, M., Qian, C., Harter, M.R., Silletti, S., Komives, E.A., Freeman, M.R., Murali, R.(2024) Nat Commun 15: 9037-9037
- PubMed: 39426953
- DOI: https://doi.org/10.1038/s41467-024-53159-8
- Primary Citation of Related Structures:
8T0F, 8T11 - PubMed Abstract:
The CUT and homeodomain are ubiquitous DNA binding elements often tandemly arranged in multiple transcription factor families. However, how the CUT and homeodomain work concertedly to bind DNA remains unknown. Using ONECUT2, a driver and therapeutic target of advanced prostate cancer, we show that while the CUT initiates DNA binding, the homeodomain thermodynamically stabilizes the ONECUT2-DNA complex through allosteric modulation of CUT. We identify an arginine pair in the ONECUT family homeodomain that can adapt to DNA sequence variations. Base interactions by this ONECUT family-specific arginine pair as well as the evolutionarily conserved residues are critical for optimal DNA binding and ONECUT2 transcriptional activity in a prostate cancer model. The evolutionarily conserved base interactions additionally determine the ONECUT2-DNA binding energetics. These findings provide insights into the cooperative DNA binding by CUT-homeodomain proteins.
- Department of Biomedical Sciences, Research Division of Immunology, Cedars-Sinai Medical Center, Los Angeles, CA, USA.
Organizational Affiliation: