Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Zhou, X., Kong, S., Maker, A., Remesh, S.G., Leung, K.K., Verba, K.A., Wells, J.A.(2024) Nat Chem Biol 20: 742-750
- PubMed: 38308046
- DOI: https://doi.org/10.1038/s41589-023-01535-8
- Primary Citation of Related Structures:
8SMK, 8SML - PubMed Abstract:
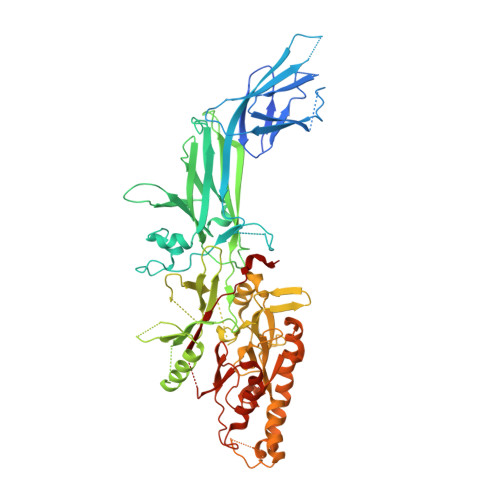
Unlocking the potential of protein arginine deiminase 4 (PAD4) as a drug target for rheumatoid arthritis requires a deeper understanding of its regulation. In this study, we use unbiased antibody selections to identify functional antibodies capable of either activating or inhibiting PAD4 activity. Through cryogenic-electron microscopy, we characterized the structures of these antibodies in complex with PAD4 and revealed insights into their mechanisms of action. Rather than steric occlusion of the substrate-binding catalytic pocket, the antibodies modulate PAD4 activity through interactions with allosteric binding sites adjacent to the catalytic pocket. These binding events lead to either alteration of the active site conformation or the enzyme oligomeric state, resulting in modulation of PAD4 activity. Our study uses antibody engineering to reveal new mechanisms for enzyme regulation and highlights the potential of using PAD4 agonist and antagonist antibodies for studying PAD4-dependency in disease models and future therapeutic development.
- Department of Pharmaceutical Chemistry, University of California San Francisco, San Francisco, CA, USA.
Organizational Affiliation: