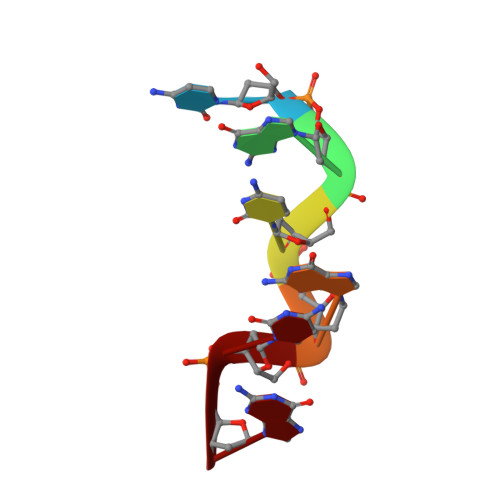Structure determination of a DNA crystal by MicroED.
Haymaker, A., Bardin, A.A., Gonen, T., Martynowycz, M.W., Nannenga, B.L.(2023) Structure 31: 1499-1503.e2
- PubMed: 37541248
- DOI: https://doi.org/10.1016/j.str.2023.07.005
- Primary Citation of Related Structures:
8SKW - PubMed Abstract:
Microcrystal electron diffraction (MicroED) is a powerful tool for determining high-resolution structures of microcrystals from a diverse array of biomolecular, chemical, and material samples. In this study, we apply MicroED to DNA crystals, which have not been previously analyzed using this technique. We utilized the d(CGCGCG) 2 DNA duplex as a model sample and employed cryo-FIB milling to create thin lamella for diffraction data collection. The MicroED data collection and subsequent processing resulted in a 1.10 Å resolution structure of the d(CGCGCG) 2 DNA, demonstrating the successful application of cryo-FIB milling and MicroED to the investigation of nucleic acid crystals.
- Biodesign Center for Applied Structural Discovery, Biodesign Institute, Arizona State University, 727 East Tyler Street, Tempe, AZ 85287, USA; School for Engineering of Matter, Transport and Energy, Arizona State University, Tempe, AZ, USA.
Organizational Affiliation: