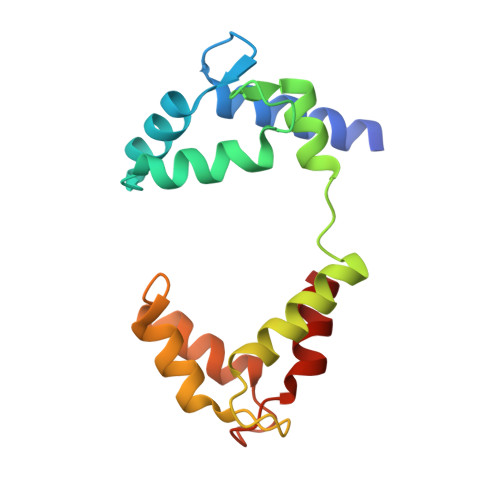
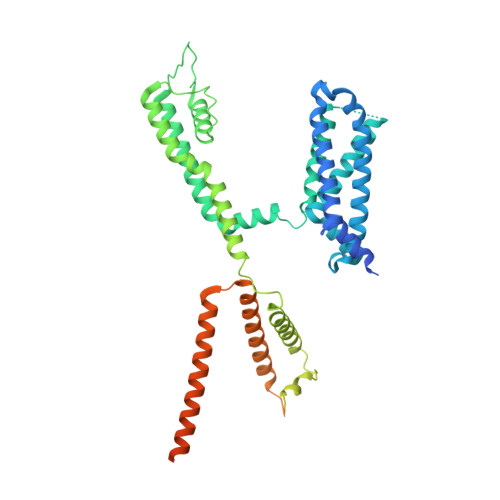
The membrane electric field regulates the PIP 2 -binding site to gate the KCNQ1 channel.
Mandala, V.S., MacKinnon, R.(2023) Proc Natl Acad Sci U S A 120: e2301985120-e2301985120
- PubMed: 37192161
- DOI: https://doi.org/10.1073/pnas.2301985120
- Primary Citation of Related Structures:
8SIK, 8SIM, 8SIN - PubMed Abstract:
Voltage-dependent ion channels underlie the propagation of action potentials and other forms of electrical activity in cells. In these proteins, voltage sensor domains (VSDs) regulate opening and closing of the pore through the displacement of their positive-charged S4 helix in response to the membrane voltage. The movement of S4 at hyperpolarizing membrane voltages in some channels is thought to directly clamp the pore shut through the S4-S5 linker helix. The KCNQ1 channel (also known as K v 7.1), which is important for heart rhythm, is regulated not only by membrane voltage but also by the signaling lipid phosphatidylinositol 4,5-bisphosphate (PIP 2 ). KCNQ1 requires PIP 2 to open and to couple the movement of S4 in the VSD to the pore. To understand the mechanism of this voltage regulation, we use cryogenic electron microscopy to visualize the movement of S4 in the human KCNQ1 channel in lipid membrane vesicles with a voltage difference across the membrane, i.e., an applied electric field in the membrane. Hyperpolarizing voltages displace S4 in such a manner as to sterically occlude the PIP 2 -binding site. Thus, in KCNQ1, the voltage sensor acts primarily as a regulator of PIP 2 binding. The voltage sensors' influence on the channel's gate is indirect through the reaction sequence: voltage sensor movement → alter PIP 2 ligand affinity → alter pore opening.
- Laboratory of Molecular Neurobiology and Biophysics, The Rockefeller University, New York, NY 10065.
Organizational Affiliation: