Semi-invariant human type II Natural Killer T cells recognise CD1d independently of bound lipids.
Chan Yew Poa, K.T.O., Le Nours, J., Rossjohn, J., Godfrey, D.G., Almeida, C.D.S., Pellicci, D.G., Moody, B.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Antigen-presenting glycoprotein CD1d | 274 | Homo sapiens | Mutation(s): 0 Gene Names: CD1D | 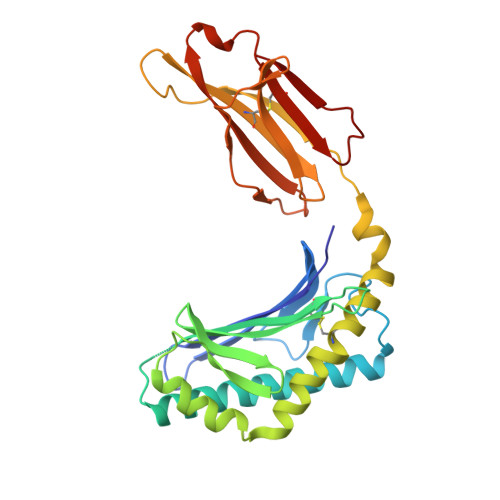 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P15813 (Homo sapiens) Explore P15813 Go to UniProtKB: P15813 | |||||
PHAROS: P15813 GTEx: ENSG00000158473 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P15813 | ||||
Glycosylation | |||||
| Glycosylation Sites: 3 | Go to GlyGen: P15813-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-2-microglobulin | 100 | Homo sapiens | Mutation(s): 0 Gene Names: B2M, CDABP0092, HDCMA22P | 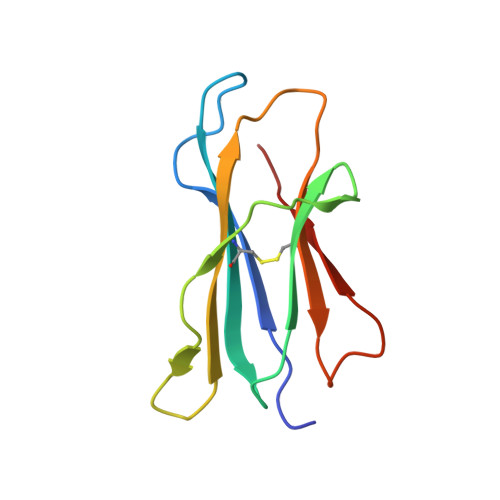 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61769 (Homo sapiens) Explore P61769 Go to UniProtKB: P61769 | |||||
PHAROS: P61769 GTEx: ENSG00000166710 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61769 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Natural killer T cell receptor TRAV26-2 | 207 | Homo sapiens | Mutation(s): 0 | 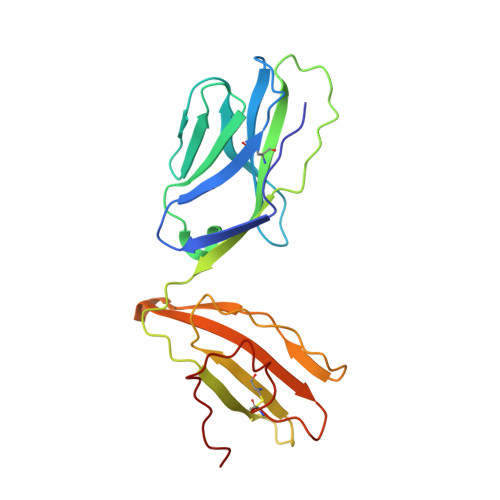 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Natural killer T cell receptor TRBV19 beta chain | 243 | Homo sapiens | Mutation(s): 0 | 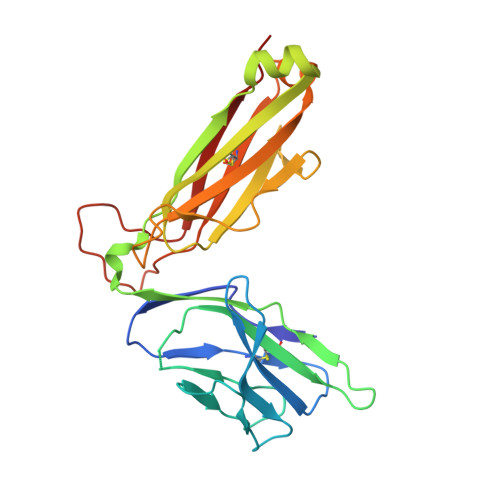 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1AHF (Subject of Investigation/LOI) Query on A1AHF | I [auth A] | N-[(2S)-3-oxo-1-(beta-L-talopyranosyloxy)octadecan-2-yl]docosanamide C46 H89 N O8 HKCYWBHIRZDQLD-HMGLXMNBSA-N |  | ||
| NAG Query on NAG | G [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| SO4 Query on SO4 | J [auth A], K [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| GOL Query on GOL | H [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 133.788 | α = 90 |
| b = 133.788 | β = 90 |
| c = 68.574 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Health and Medical Research Council (NHMRC, Australia) | Australia | -- |