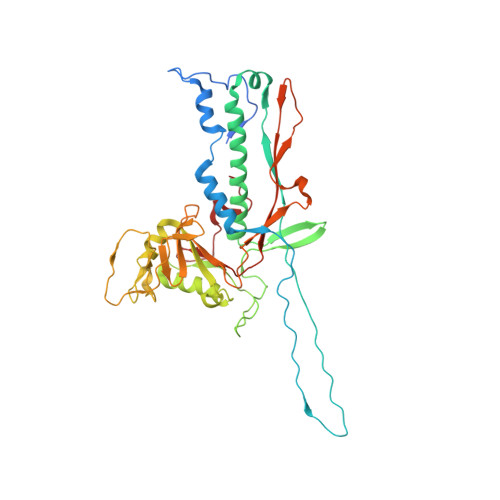
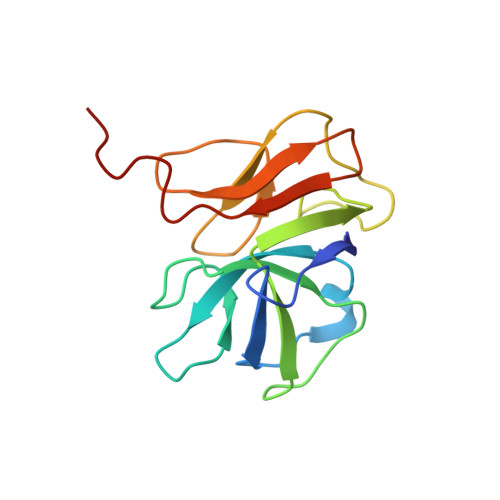
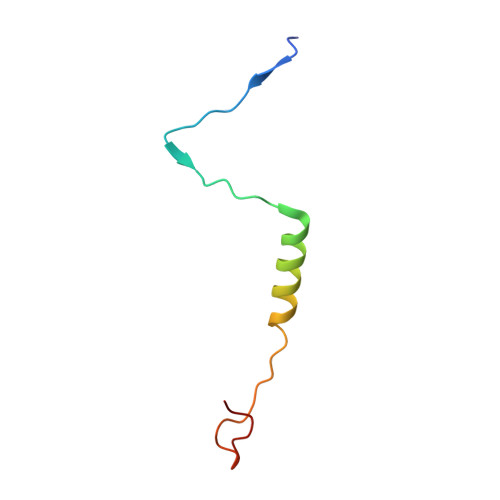
Stabilization mechanism accommodating genome length variation in evolutionarily related viral capsids.
Podgorski, J.M., Podgorski, J., Abad, L., Jacobs-Sera, D., Freeman, K.G., Brown, C., Hatfull, G.F., Luque, A., White, S.J.(2025) Nat Commun 16: 3145-3145
- PubMed: 40175362
- DOI: https://doi.org/10.1038/s41467-025-58298-0
- Primary Citation of Related Structures:
8SAJ - PubMed Abstract:
Tailed bacteriophages are one of the most numerous and diverse group of viruses. They store their genome at quasi-crystalline densities in capsids built from multiple copies of proteins adopting the HK97-fold. The high density of the genome exerts an internal pressure, requiring a maturation process that reinforces their capsids. However, it is unclear how capsid stabilization strategies have adapted to accommodate the evolution of larger genomes in this virus group. Here we characterize a capsid reinforcement mechanism in two evolutionary-related actinobacteriophages that modifies the length of a stabilization protein to accommodate a larger genome while maintaining the same capsid size. We use cryo-EM to reveal that capsids contain split hexamers of HK97-fold proteins with a stabilization protein in the chasm. The observation of split hexamers in mature capsids is unprecedented, so we rationalize this result mathematically, discovering that icosahedral capsids can be formed by all split or skewed hexamers as long as their T-number is not a multiple of three. Our results suggest that analogous stabilization mechanisms can be present in other icosahedral capsids, and they provide a strategy for engineering capsids accommodating larger DNA cargoes as gene delivery systems.
- Biology/Physics Building, Department of Molecular and Cell Biology, University of Connecticut, 91 North Eagleville Road, Unit-3125, Storrs, CT, USA.
Organizational Affiliation: