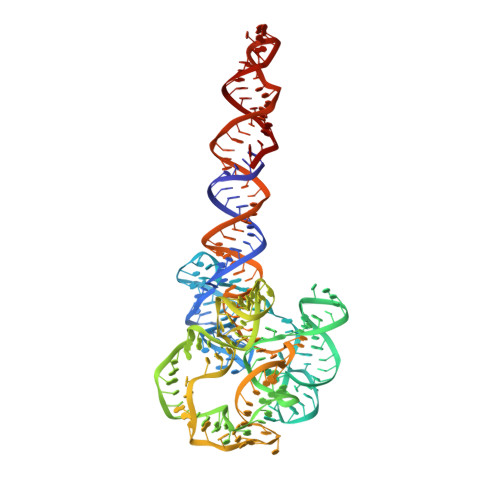
Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Ding, J., Deme, J.C., Stagno, J.R., Yu, P., Lea, S.M., Wang, Y.X.(2023) Nucleic Acids Res 51: 9952-9960
- PubMed: 37534568
- DOI: https://doi.org/10.1093/nar/gkad651
- Primary Citation of Related Structures:
8SA2, 8SA3, 8SA4, 8SA5, 8SA6 - PubMed Abstract:
RNA conformational heterogeneity often hampers its high-resolution structure determination, especially for large and flexible RNAs devoid of stabilizing proteins or ligands. The adenosylcobalamin riboswitch exhibits heterogeneous conformations under 1 mM Mg2+ concentration and ligand binding reduces conformational flexibility. Among all conformers, we determined one apo (5.3 Å) and four holo cryo-electron microscopy structures (overall 3.0-3.5 Å, binding pocket 2.9-3.2 Å). The holo dimers exhibit global motions of helical twisting and bending around the dimer interface. A backbone comparison of the apo and holo states reveals a large structural difference in the P6 extension position. The central strand of the binding pocket, junction 6/3, changes from an 'S'- to a 'U'-shaped conformation to accommodate ligand. Furthermore, the binding pocket can partially form under 1 mM Mg2+ and fully form under 10 mM Mg2+ within the bound-like structure in the absence of ligand. Our results not only demonstrate the stabilizing ligand-induced conformational changes in and around the binding pocket but may also provide further insight into the role of the P6 extension in ligand binding and selectivity.
- Protein-Nucleic Acid Interaction Section, Center for Structural Biology, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.
Organizational Affiliation: