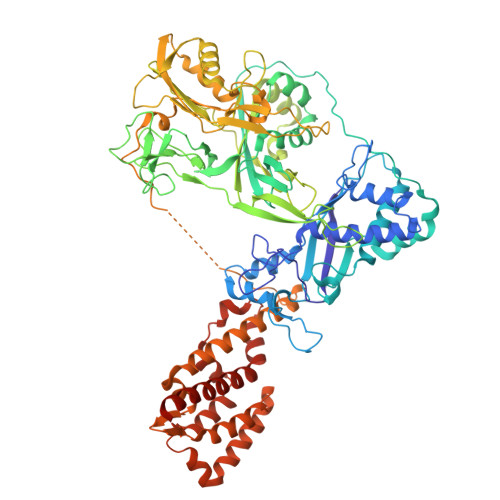
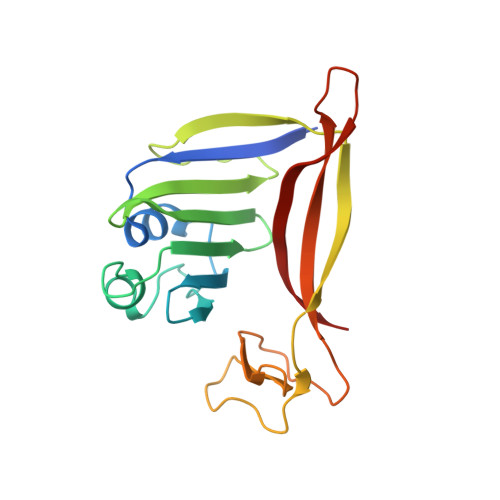
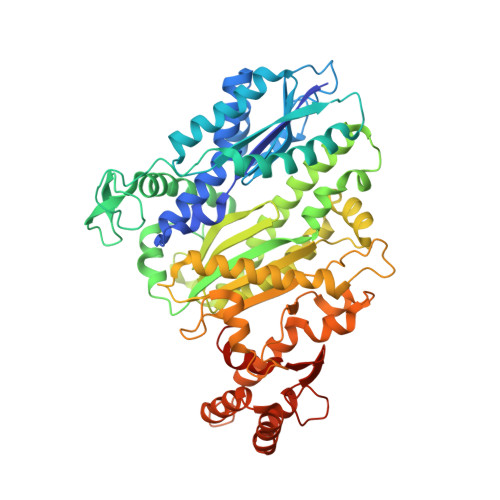
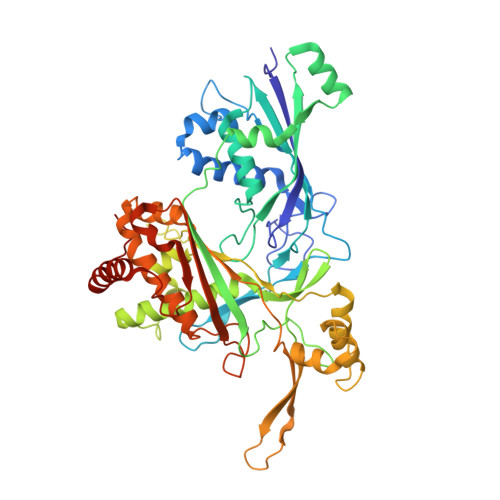
RNA targeting and cleavage by the type III-Dv CRISPR effector complex.
Schwartz, E.A., Bravo, J.P.K., Ahsan, M., Macias, L.A., McCafferty, C.L., Dangerfield, T.L., Walker, J.N., Brodbelt, J.S., Palermo, G., Fineran, P.C., Fagerlund, R.D., Taylor, D.W.(2024) Nat Commun 15: 3324-3324
- PubMed: 38637512
- DOI: https://doi.org/10.1038/s41467-024-47506-y
- Primary Citation of Related Structures:
8S9T, 8S9U, 8S9V, 8S9X - PubMed Abstract:
CRISPR-Cas are adaptive immune systems in bacteria and archaea that utilize CRISPR RNA-guided surveillance complexes to target complementary RNA or DNA for destruction 1-5 . Target RNA cleavage at regular intervals is characteristic of type III effector complexes 6-8 . Here, we determine the structures of the Synechocystis type III-Dv complex, an apparent evolutionary intermediate from multi-protein to single-protein type III effectors 9,10 , in pre- and post-cleavage states. The structures show how multi-subunit fusion proteins in the effector are tethered together in an unusual arrangement to assemble into an active and programmable RNA endonuclease and how the effector utilizes a distinct mechanism for target RNA seeding from other type III effectors. Using structural, biochemical, and quantum/classical molecular dynamics simulation, we study the structure and dynamics of the three catalytic sites, where a 2'-OH of the ribose on the target RNA acts as a nucleophile for in line self-cleavage of the upstream scissile phosphate. Strikingly, the arrangement at the catalytic residues of most type III complexes resembles the active site of ribozymes, including the hammerhead, pistol, and Varkud satellite ribozymes. Our work provides detailed molecular insight into the mechanisms of RNA targeting and cleavage by an important intermediate in the evolution of type III effector complexes.
- Interdisciplinary Life Sciences Graduate Programs, University of Texas at Austin, Austin, TX, USA.
Organizational Affiliation: