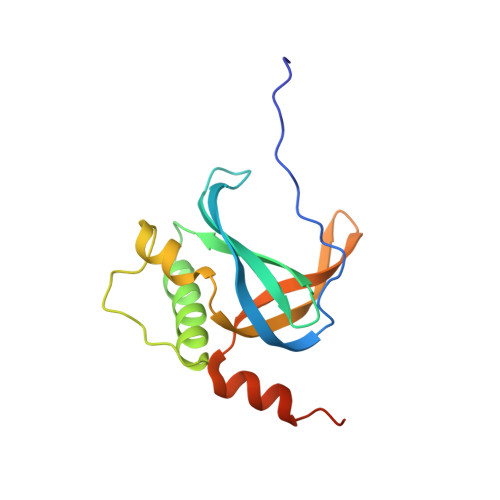
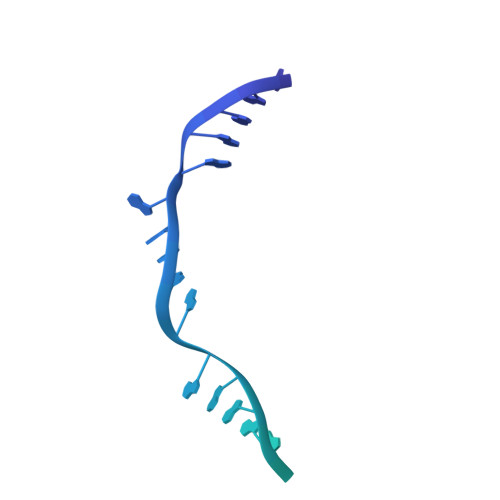
PrgE: an OB-fold protein from plasmid pCF10 with striking differences to prototypical bacterial SSBs.
Breidenstein, A., Lamy, A., Bader, C.P., Sun, W.S., Wanrooij, P.H., Berntsson, R.P.(2024) Life Sci Alliance 7
- PubMed: 38811160
- DOI: https://doi.org/10.26508/lsa.202402693
- Primary Citation of Related Structures:
8S4S, 8S4T - PubMed Abstract:
A major pathway for horizontal gene transfer is the transmission of DNA from donor to recipient cells via plasmid-encoded type IV secretion systems (T4SSs). Many conjugative plasmids encode for a single-stranded DNA-binding protein (SSB) together with their T4SS. Some of these SSBs have been suggested to aid in establishing the plasmid in the recipient cell, but for many, their function remains unclear. Here, we characterize PrgE, a proposed SSB from the Enterococcus faecalis plasmid pCF10. We show that PrgE is not essential for conjugation. Structurally, it has the characteristic OB-fold of SSBs, but it has very unusual DNA-binding properties. Our DNA-bound structure shows that PrgE binds ssDNA like beads on a string supported by its N-terminal tail. In vitro studies highlight the plasticity of PrgE oligomerization and confirm the importance of the N-terminus. Unlike other SSBs, PrgE binds both double- and single-stranded DNA equally well. This shows that PrgE has a quaternary assembly and DNA-binding properties that are very different from the prototypical bacterial SSB, but also different from eukaryotic SSBs.
- https://ror.org/05kb8h459 Department of Medical Biochemistry and Biophysics, Umeå University, Umeå, Sweden.
Organizational Affiliation: