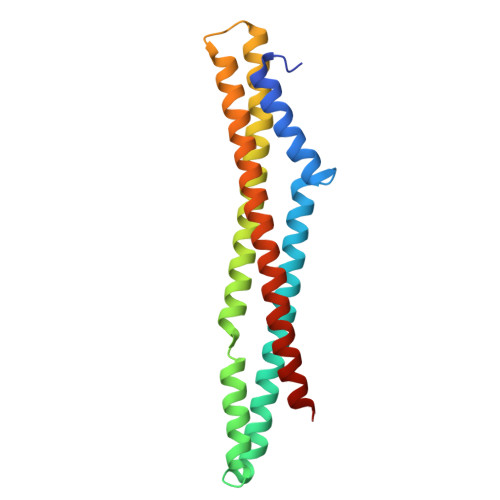
Divergent dimerization mechanisms and conserved DNA-binding function in PFam12 proteins of Borrelia burgdorferi.
Brangulis, K., Tupina, D., Sinicina, E.E., Zelencova-Gopejenko, D., Akopjana, I., Bogans, J., Tars, K.(2025) Sci Rep 15: 11518-11518
- PubMed: 40181002
- DOI: https://doi.org/10.1038/s41598-025-93944-z
- Primary Citation of Related Structures:
8S2F, 8S2P - PubMed Abstract:
Lyme disease, caused by the spirochete Borrelia burgdorferi, is transmitted to mammalian hosts during the feeding process of infected Ixodes ticks. Our previous studies demonstrated that the paralogous gene family 12 (PFam12) consisting of five members (BBK01, BBG01, BBH37, BBJ08, and BB0844) are non-specific DNA-binding proteins. PFam12 proteins share 31-69% sequence identity, are located either on the surface or within the periplasm and are upregulated as the tick starts its blood meal. The crystal structure of BBK01 revealed that the protein forms a homodimer, which is potentially critical for DNA binding. In this study, we determined the crystal structure of another PFam12 member, BBH37, to gain a better insight into this unique paralogous family. Although BBK01 dimerization is mediated by its C-terminal region and is thought to be critical for DNA binding, BBH37 forms dimers through an alternative mechanism where a unique disulfide bond is involved. We found that BBH37 is still able to interact with DNA with micromolar affinity. Molecular dynamics simulations and site-directed mutagenesis was conducted to characterize these unique DNA binding proteins. This study highlights the structural diversity within the PFam12, demonstrating that despite significant differences in dimerization mechanisms, these proteins retain their DNA-binding capability.
- Latvian Biomedical Research and Study Centre, Ratsupites 1 K-1, Riga, 1067, Latvia. kalvis@biomed.lu.lv.
Organizational Affiliation: