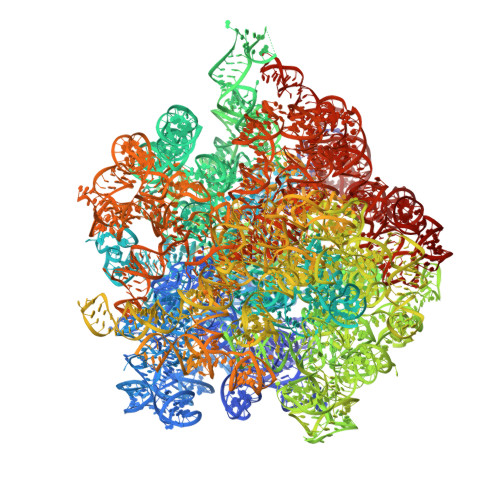
A role for the S4-domain containing protein YlmH in ribosome-associated quality control in Bacillus subtilis.
Takada, H., Paternoga, H., Fujiwara, K., Nakamoto, J.A., Park, E.N., Dimitrova-Paternoga, L., Beckert, B., Saarma, M., Tenson, T., Buskirk, A.R., Atkinson, G.C., Chiba, S., Wilson, D.N., Hauryliuk, V.(2024) Nucleic Acids Res 52: 8483-8499
- PubMed: 38811035
- DOI: https://doi.org/10.1093/nar/gkae399
- Primary Citation of Related Structures:
8S1P, 8S1U - PubMed Abstract:
Ribosomes trapped on mRNAs during protein synthesis need to be rescued for the cell to survive. The most ubiquitous bacterial ribosome rescue pathway is trans-translation mediated by tmRNA and SmpB. Genetic inactivation of trans-translation can be lethal, unless ribosomes are rescued by ArfA or ArfB alternative rescue factors or the ribosome-associated quality control (RQC) system, which in Bacillus subtilis involves MutS2, RqcH, RqcP and Pth. Using transposon sequencing in a trans-translation-incompetent B. subtilis strain we identify a poorly characterized S4-domain-containing protein YlmH as a novel potential RQC factor. Cryo-EM structures reveal that YlmH binds peptidyl-tRNA-50S complexes in a position analogous to that of S4-domain-containing protein RqcP, and that, similarly to RqcP, YlmH can co-habit with RqcH. Consistently, we show that YlmH can assume the role of RqcP in RQC by facilitating the addition of poly-alanine tails to truncated nascent polypeptides. While in B. subtilis the function of YlmH is redundant with RqcP, our taxonomic analysis reveals that in multiple bacterial phyla RqcP is absent, while YlmH and RqcH are present, suggesting that in these species YlmH plays a central role in the RQC.
- Faculty of Life Sciences, Kyoto Sangyo University and Institute for Protein Dynamics, Kamigamo, Motoyama, Kita-ku, Kyoto 603-8555, Japan.
Organizational Affiliation: