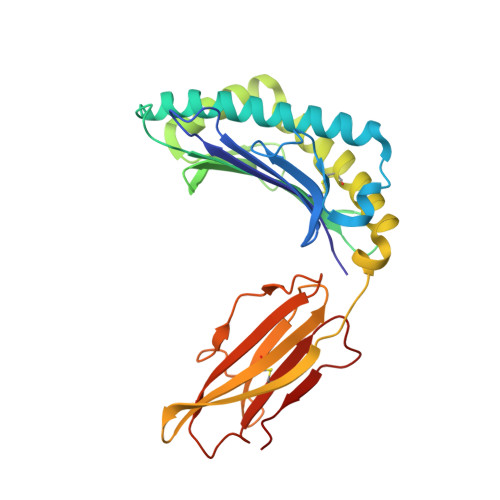
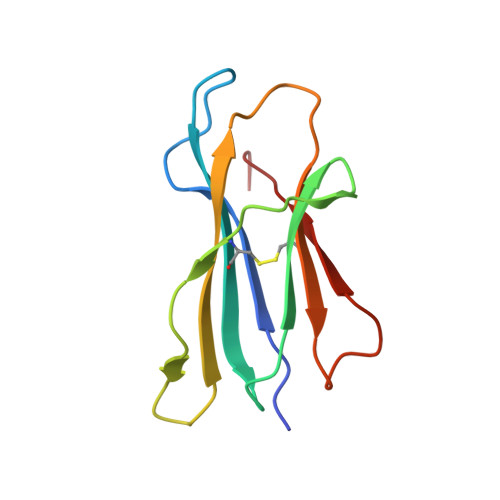
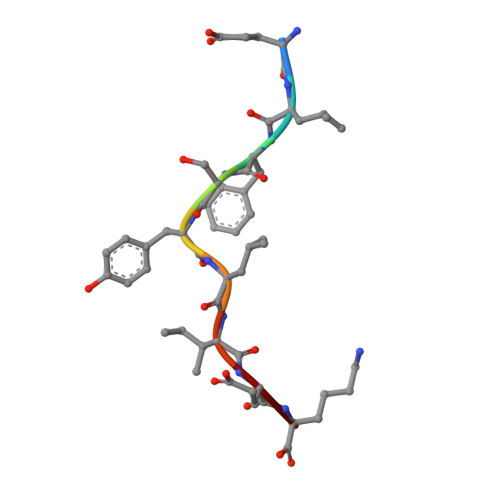
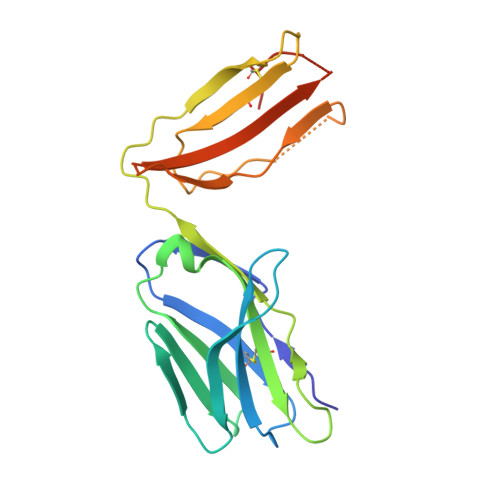
Broadening alloselectivity of T cell receptors by structure guided engineering.
Karuppiah, V., Sangani, D., Whaley, L., Pengelly, R., Uluocak, P., Carreira, R.J., Hock, M., Cristina, P.D., Bartasun, P., Dobrinic, P., Smith, N., Barnbrook, K., Robinson, R.A., Harper, S.(2024) Sci Rep 14: 26851-26851
- PubMed: 39500929
- DOI: https://doi.org/10.1038/s41598-024-75140-7
- Primary Citation of Related Structures:
8RYM, 8RYN, 8RYO, 8RYP, 8RYQ - PubMed Abstract:
Specificity of a T cell receptor (TCR) is determined by the combination of its interactions to the peptide and human leukocyte antigen (HLA). TCR-based therapeutic molecules have to date targeted a single peptide in the context of a single HLA allele. Some peptides are presented on multiple HLA alleles, and by engineering TCRs for specific recognition of more than one allele, there is potential to expand the targetable patient population. Here, as a proof of concept, we studied two TCRs, S2 and S8, binding to the PRAME peptide antigen (ELFSYLIEK) presented by HLA alleles HLA-A*03:01 and HLA-A*11:01. By structure-guided affinity maturation targeting a specific residue on the HLA surface, we show that the affinity of the TCR can be modulated for different alleles. Using a combination of affinity maturation and functional T cell assay, we demonstrate that an engineered TCR can target the same peptide on two different HLA alleles with similar affinity and potency. This work highlights the importance of engineering alloselectivity for designing TCR based therapeutics suitable for differing global populations.
- Immunocore Ltd, 92 Milton Park, Abingdon, Oxfordshire, OX14 4RY, UK. Vijaykumar.Karuppiah@immunocore.com.
Organizational Affiliation: