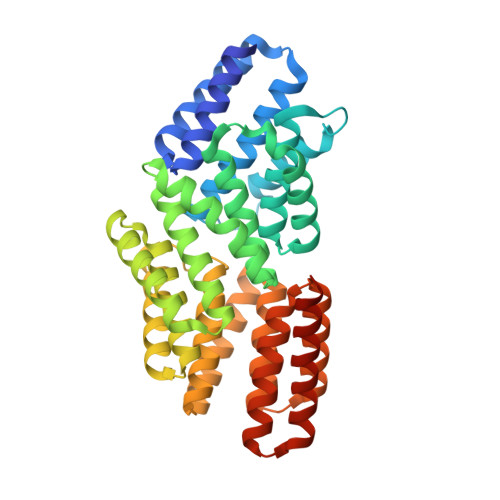
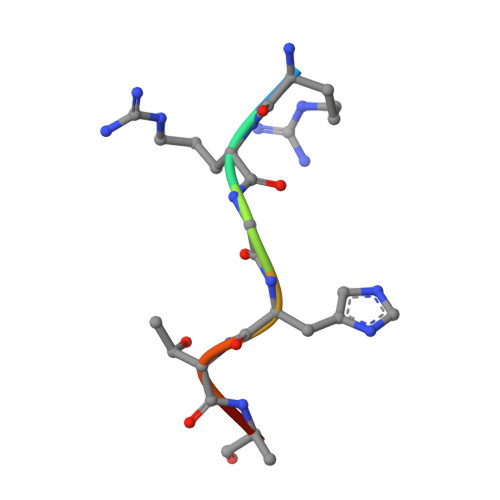
Extracellular proteolysis of tandemly duplicated pheromone propeptides affords additional complexity to bacterial quorum sensing.
Felipe-Ruiz, A., Zamora-Caballero, S., Bendori, S.O., Penades, J.R., Eldar, A., Marina, A.(2024) PLoS Biol 22: e3002744-e3002744
- PubMed: 39137235
- DOI: https://doi.org/10.1371/journal.pbio.3002744
- Primary Citation of Related Structures:
8RST, 8RSU, 8RSV, 8RTC, 8RTE - PubMed Abstract:
Bacterial interactions are vital for adapting to changing environments, with quorum sensing (QS) systems playing a central role in coordinating behaviors through small signaling molecules. The RRNPPA family is the prevalent QS systems in Bacillota and mediating communication through secreted oligopeptides, which are processed into active pheromones by extracellular proteases. Notably, in several cases the propeptides show the presence of multiple putative pheromones within their sequences, which has been proposed as a mechanism to diversify peptide-receptor specificity and potentially facilitate new functions. However, neither the processes governing the maturation of propeptides containing multiple pheromones, nor their functional significance has been evaluated. Here, using 2 Rap systems from bacteriophages infecting Bacillus subtilis that exhibit different types of pheromone duplication in their propeptides, we investigate the maturation process and the molecular and functional activities of the produced pheromones. Our results reveal that distinct maturation processes generate multiple mature pheromones, which bind to receptors with varying affinities but produce identical structural and biological responses. These findings add additional layers in the complexity of QS communication and regulation, opening new possibilities for microbial social behaviors, highlighting the intricate nature of bacterial interactions and adaptation.
Organizational Affiliation:
Instituto de Biomedicina de Valencia (IBV)-CSIC and CIBER de Enfermedades Raras (CIBERER)-ISCIII, Valencia, Spain.