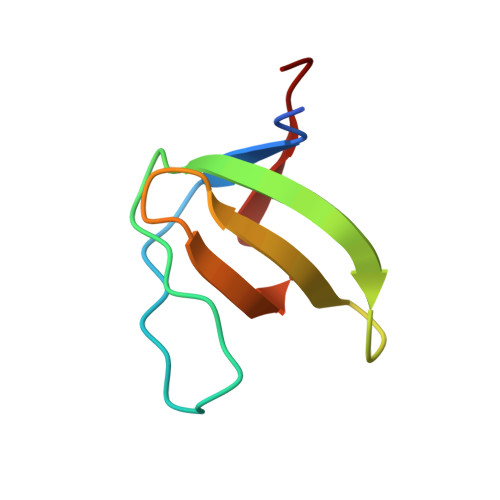
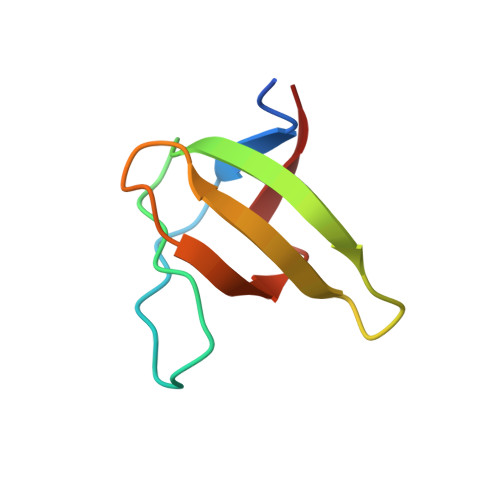
Structural basis of homodimerization of the JNK scaffold protein JIP2 and its heterodimerization with JIP1.
Marino Perez, L., Ielasi, F.S., Lee, A., Delaforge, E., Juyoux, P., Tengo, M., Davis, R.J., Palencia, A., Jensen, M.R.(2024) Structure 32: 1394
- PubMed: 39013462
- DOI: https://doi.org/10.1016/j.str.2024.06.010
- Primary Citation of Related Structures:
8RPP - PubMed Abstract:
The scaffold proteins JIP1 and JIP2 intervene in the c-Jun N-terminal kinase (JNK) pathway to mediate signaling specificity by coordinating the simultaneous assembly of multiple kinases. Using NMR, we demonstrate that JIP1 and JIP2 heterodimerize via their SH3 domains with the affinity of heterodimerization being comparable to homodimerization. We present the high-resolution crystal structure of the JIP2-SH3 homodimer and the JIP1-JIP2-SH3 heterodimeric complex. The JIP2-SH3 structure reveals how charge differences in residues at its dimer interface lead to formation of compensatory hydrogen bonds and salt bridges, distinguishing it from JIP1-SH3. In the JIP1-JIP2-SH3 complex, structural features of each homodimer are employed to stabilize the heterodimer. Building on these insights, we identify key residues crucial for stabilizing the dimer of both JIP1 and JIP2. Through targeted mutations in cellulo, we demonstrate a functional role for the dimerization of the JIP1 and JIP2 scaffold proteins in activation of the JNK signaling pathway.
- University Grenoble Alpes, CEA, CNRS, IBS, Grenoble, France; Departament de Química, Universitat de les Illes Balears, Institut Universitari d'Investigació en Ciències de la Salut (IUNICS), Institut de Recerca en Ciències de la Salut (IdISBa), Palma, Spain.
Organizational Affiliation: