The AMP-forming acetyl-CoA-synthetase is regulated by lysine acetylation and has an intrinsic phosphotransacetylase activity to adjust its activity to the cellular acetyl-phosphate level
Qin, C., Lammers, M.(2024) Nature Comm
Experimental Data Snapshot
(2024) Nature Comm
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acetate--CoA ligase | 657 | Chloroflexota bacterium | Mutation(s): 0 Gene Names: acs, E6J36_13485 EC: 6.2.1.1 | 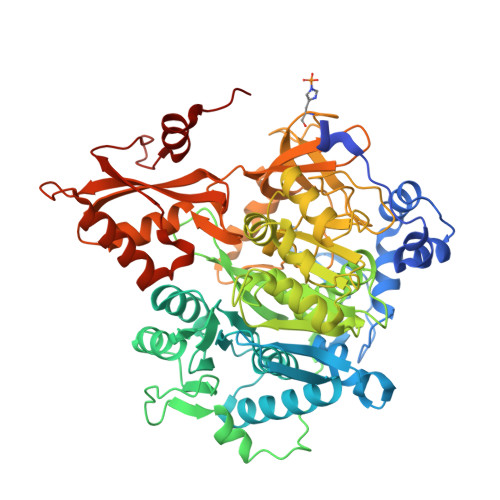 | |
UniProt | |||||
Find proteins for A0A535FEC2 (Chloroflexota bacterium) Explore A0A535FEC2 Go to UniProtKB: A0A535FEC2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A535FEC2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 6R9 (Subject of Investigation/LOI) Query on 6R9 | E [auth A], H [auth B], K [auth C], N [auth D] | [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] ethanoate C12 H16 N5 O8 P UBPVOHPZRZIJHM-WOUKDFQISA-N |  | ||
| EPE Query on EPE | F [auth A], I [auth B], L [auth C], O [auth D] | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID C8 H18 N2 O4 S JKMHFZQWWAIEOD-UHFFFAOYSA-N |  | ||
| MG Query on MG | G [auth A], J [auth B], M [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA Query on NA | P [auth D] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| NEP Query on NEP | A, B, C, D | L-PEPTIDE LINKING | C6 H10 N3 O5 P |  | HIS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 161.909 | α = 90 |
| b = 161.909 | β = 90 |
| c = 817.234 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |