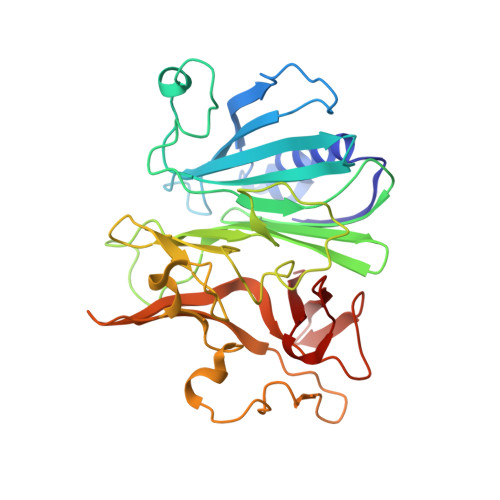
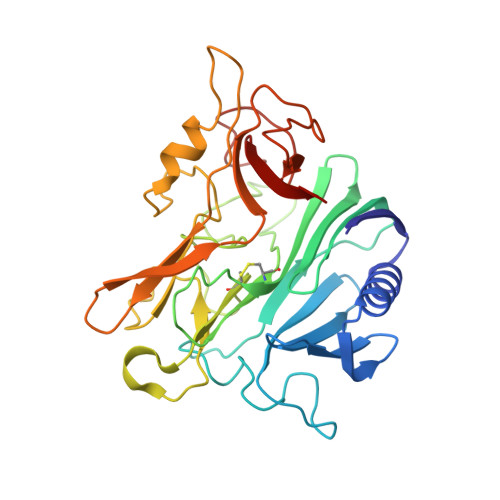
Structure and replication of Pseudomonas aeruginosa phage JBD30.
Valentova, L., Fuzik, T., Novacek, J., Hlavenkova, Z., Pospisil, J., Plevka, P.(2024) EMBO J 43: 4384-4405
- PubMed: 39143239
- DOI: https://doi.org/10.1038/s44318-024-00195-1
- Primary Citation of Related Structures:
8RK3, 8RK4, 8RK5, 8RK6, 8RK7, 8RK8, 8RK9, 8RKA, 8RKB, 8RKC, 8RKN, 8RKO, 8RKX, 8RQE - PubMed Abstract:
Bacteriophages are the most abundant biological entities on Earth, but our understanding of many aspects of their lifecycles is still incomplete. Here, we have structurally analysed the infection cycle of the siphophage Casadabanvirus JBD30. Using its baseplate, JBD30 attaches to Pseudomonas aeruginosa via the bacterial type IV pilus, whose subsequent retraction brings the phage to the bacterial cell surface. Cryo-electron microscopy structures of the baseplate-pilus complex show that the tripod of baseplate receptor-binding proteins attaches to the outer bacterial membrane. The tripod and baseplate then open to release three copies of the tape-measure protein, an event that is followed by DNA ejection. JBD30 major capsid proteins assemble into procapsids, which expand by 7% in diameter upon filling with phage dsDNA. The DNA-filled heads are finally joined with 180-nm-long tails, which bend easily because flexible loops mediate contacts between the successive discs of major tail proteins. It is likely that the structural features and replication mechanisms described here are conserved among siphophages that utilize the type IV pili for initial cell attachment.
- Central European Institute of Technology, Masaryk University, Brno, Czech Republic.
Organizational Affiliation: