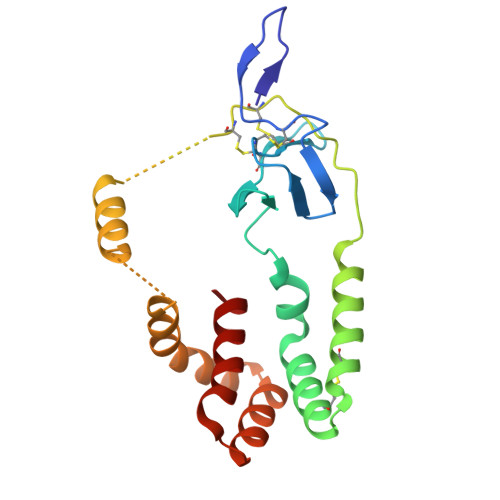
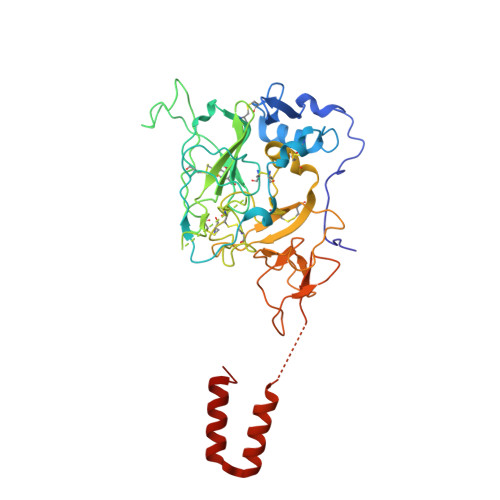
The hepatitis C virus envelope protein complex is a dimer of heterodimers.
Augestad, E.H., Holmboe Olesen, C., Gronberg, C., Soerensen, A., Velazquez-Moctezuma, R., Fanalista, M., Bukh, J., Wang, K., Gourdon, P., Prentoe, J.(2024) Nature 633: 704-709
- PubMed: 39232163
- DOI: https://doi.org/10.1038/s41586-024-07783-5
- Primary Citation of Related Structures:
8RJJ, 8RK0 - PubMed Abstract:
Fifty-eight million individuals worldwide are affected by chronic hepatitis C virus (HCV) infection, a primary driver of liver cancer for which no vaccine is available 1 . The HCV envelope proteins E1 and E2 form a heterodimer (E1/E2), which is the target for neutralizing antibodies 2 . However, the higher-order organization of these E1/E2 heterodimers, as well as that of any Hepacivirus envelope protein complex, remains unknown. Here we determined the cryo-electron microscopy structure of two E1/E2 heterodimers in a homodimeric arrangement. We reveal how the homodimer is established at the molecular level and provide insights into neutralizing antibody evasion and membrane fusion by HCV, as orchestrated by E2 motifs such as hypervariable region 1 and antigenic site 412, as well as the organization of the transmembrane helices, including two internal to E1. This study addresses long-standing questions on the higher-order oligomeric arrangement of Hepacivirus envelope proteins and provides a critical framework in the design of novel HCV vaccine antigens.
- Copenhagen Hepatitis C Program (CO-HEP), Department of Infectious Diseases, Copenhagen University Hospital, Hvidovre, Denmark. elias.augestad@sund.ku.dk.
Organizational Affiliation: