Identification of a novel inhibitor for beta-galactosidase from Pseudomonas syringae.
Hardenbrook, N.J., Sheng, Y., Sanguankiattichai, N., Zhang, P., Preston, G., van der Hoorn, R.To be published.
Experimental Data Snapshot
Starting Model: other
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-galactosidase | 1,030 | Escherichia coli | Mutation(s): 0 Gene Names: lacZ, b0344, JW0335 EC: 3.2.1.23 | 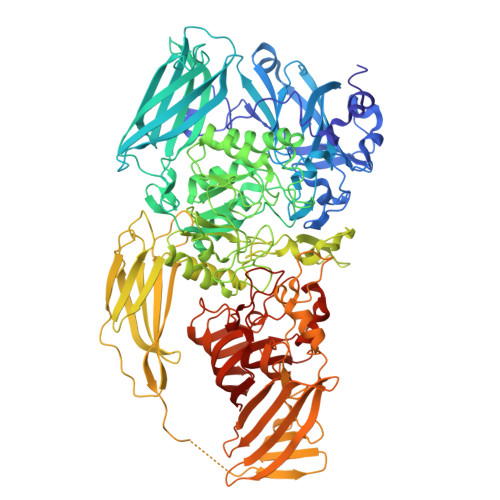 | |
UniProt | |||||
Find proteins for P00722 (Escherichia coli (strain K12)) Explore P00722 Go to UniProtKB: P00722 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00722 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1H05 (Subject of Investigation/LOI) Query on A1H05 | E [auth A], J [auth B], O [auth C], T [auth D] | (2S,3R,4S)-2-[bis(oxidanyl)methyl]pyrrolidine-3,4-diol C5 H11 N O4 GRHIWHJZSCBMFQ-HZLVTQRSSA-N |  | ||
| MG Query on MG | F [auth A] G [auth A] K [auth B] L [auth B] P [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA Query on NA | H [auth A] I [auth A] M [auth B] N [auth B] R [auth C] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.19.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/T015128/1 |
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/R017913/1 |
| European Research Council (ERC) | European Union | 101021133 |
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | U54 AI170791-7522 |
| Wellcome Trust | United Kingdom | 206422/Z/17/Z |