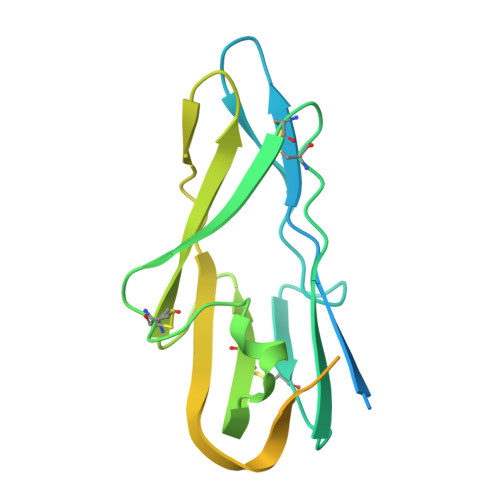
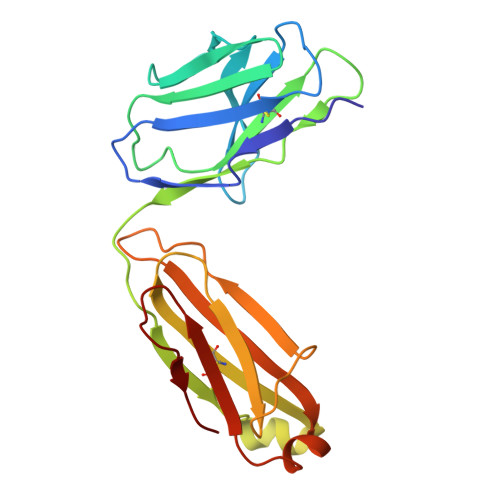
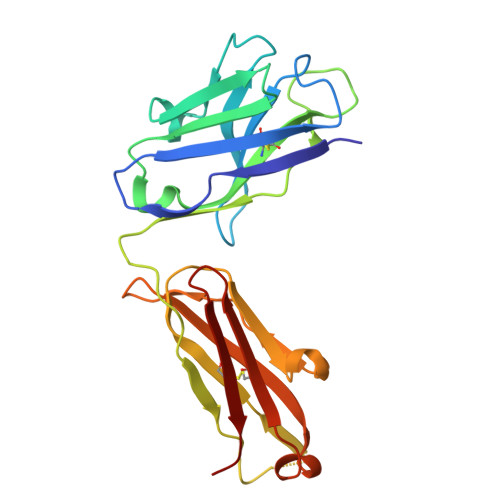
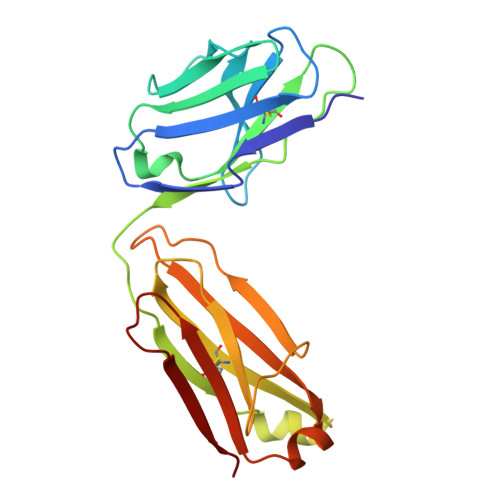
Neutralizing antibodies reveal cryptic vulnerabilities and interdomain crosstalk in the porcine deltacoronavirus spike protein.
Du, W., Debski-Antoniak, O., Drabek, D., van Haperen, R., van Dortmondt, M., van der Lee, J., Drulyte, I., van Kuppeveld, F.J.M., Grosveld, F., Hurdiss, D.L., Bosch, B.J.(2024) Nat Commun 15: 5330-5330
- PubMed: 38909062
- DOI: https://doi.org/10.1038/s41467-024-49693-0
- Primary Citation of Related Structures:
8R9W, 8R9X, 8R9Y, 8R9Z - PubMed Abstract:
Porcine deltacoronavirus (PDCoV) is an emerging enteric pathogen that has recently been detected in humans. Despite this zoonotic concern, the antigenic structure of PDCoV remains unknown. The virus relies on its spike (S) protein for cell entry, making it a prime target for neutralizing antibodies. Here, we generate and characterize a set of neutralizing antibodies targeting the S protein, shedding light on PDCoV S interdomain crosstalk and its vulnerable sites. Among the four identified antibodies, one targets the S1A domain, causing local and long-range conformational changes, resulting in partial exposure of the S1B domain. The other antibodies bind the S1B domain, disrupting binding to aminopeptidase N (APN), the entry receptor for PDCoV. Notably, the epitopes of these S1B-targeting antibodies are concealed in the prefusion S trimer conformation, highlighting the necessity for conformational changes for effective antibody binding. The binding footprint of one S1B binder entirely overlaps with APN-interacting residues and thus targets a highly conserved epitope. These findings provide structural insights into the humoral immune response against the PDCoV S protein, potentially guiding vaccine and therapeutic development for this zoonotic pathogen.
- Virology Section, Infectious Diseases and Immunology Division, Department of Biomolecular Health Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Organizational Affiliation: