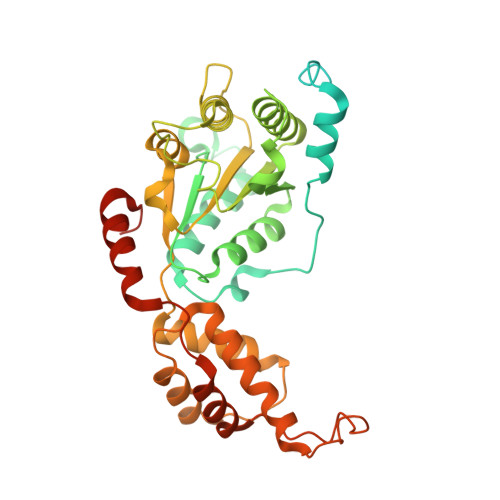
Molecular basis of FIGNL1 in dissociating RAD51 from DNA and chromatin.
Carver, A., Yu, T.Y., Yates, L.A., White, T., Wang, R., Lister, K., Jasin, M., Zhang, X.(2025) Science 387: 426-431
- PubMed: 39636933
- DOI: https://doi.org/10.1126/science.adr7920
- Primary Citation of Related Structures:
8R64 - PubMed Abstract:
Maintaining genome integrity is an essential and challenging process. RAD51 recombinase, the central player of several crucial processes in repairing DNA and protecting genome integrity, forms filaments on DNA, which are tightly regulated. One of these RAD51 regulators is FIGNL1, that prevents persistent RAD51 foci without or after DNA damage and genotoxic chromatin association in cells. The cryogenic electron microscopy structure of FIGNL1 in complex with RAD51 reveals that FIGNL1 forms a non-planar hexamer and RAD51 N terminus enclosure in the FIGNL1 hexamer pore. Mutations in pore loop or catalytic residues of FIGNL1 render it defective in filament disassembly and are lethal in mouse embryonic stem cells. Our study reveals a unique mechanism for removing RAD51 from bound substrates and provides the molecular basis for FIGNL1 in maintaining genome stability.
- DNA Processing Machines Laboratory, Francis Crick Institute, London, UK.
Organizational Affiliation: