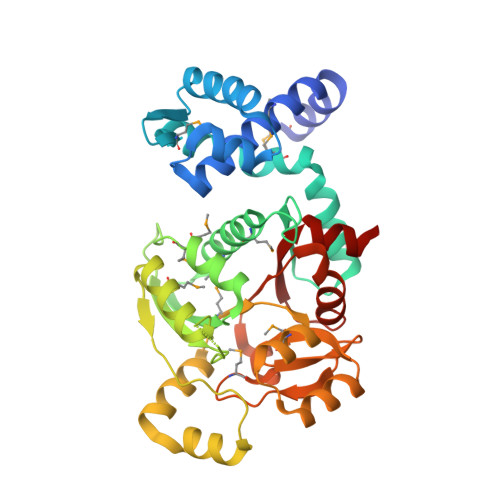
Structural characterization of two prototypical repressors of SorC family reveals tetrameric assemblies on DNA and mechanism of function.
Soltysova, M., Skerlova, J., Pachl, P., Skubnik, K., Fabry, M., Sieglova, I., Farolfi, M., Grishkovskaya, I., Babiak, M., Novacek, J., Krasny, L., Rezacova, P.(2024) Nucleic Acids Res 52: 7305-7320
- PubMed: 38842936
- DOI: https://doi.org/10.1093/nar/gkae434
- Primary Citation of Related Structures:
8R3G, 8R7Y - PubMed Abstract:
The SorC family of transcriptional regulators plays a crucial role in controlling the carbohydrate metabolism and quorum sensing. We employed an integrative approach combining X-ray crystallography and cryo-electron microscopy to investigate architecture and functional mechanism of two prototypical representatives of two sub-classes of the SorC family: DeoR and CggR from Bacillus subtilis. Despite possessing distinct DNA-binding domains, both proteins form similar tetrameric assemblies when bound to their respective DNA operators. Structural analysis elucidates the process by which the CggR-regulated gapA operon is derepressed through the action of two effectors: fructose-1,6-bisphosphate and newly confirmed dihydroxyacetone phosphate. Our findings provide the first comprehensive understanding of the DNA binding mechanism of the SorC-family proteins, shedding new light on their functional characteristics.
- Structural Biology, Institute of Organic Chemistry and Biochemistry of Czech Academy of Sciences, Prague, 166 10, Czechia.
Organizational Affiliation: