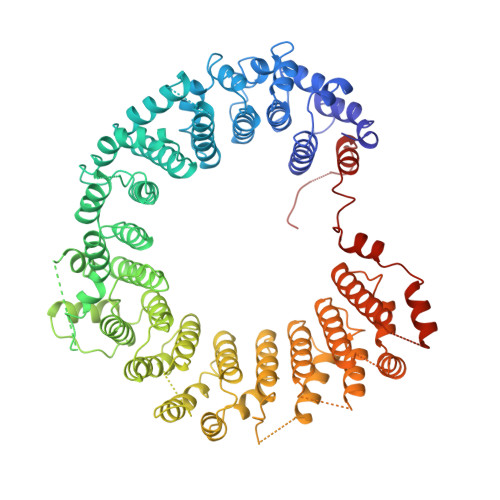
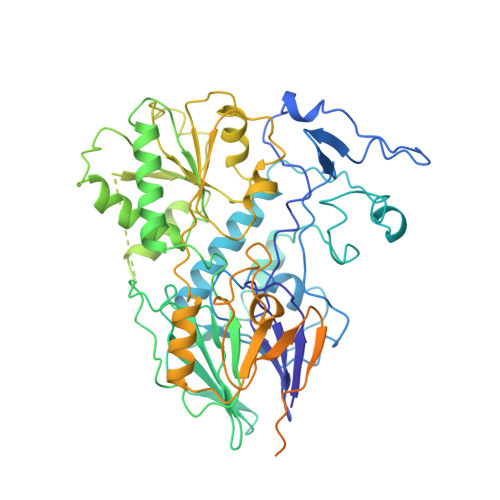
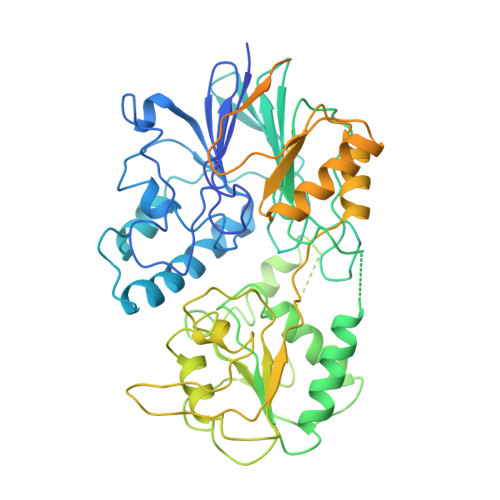
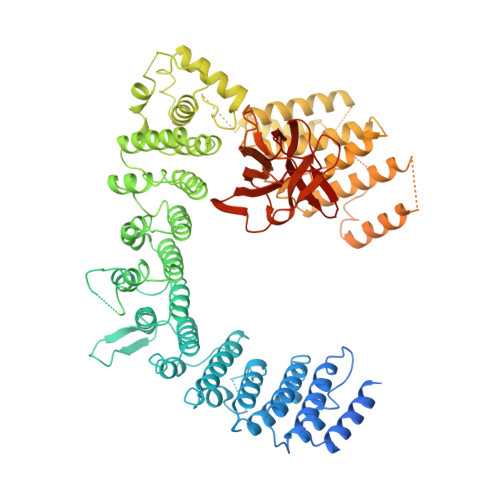
Assembly mechanism of Integrator's RNA cleavage module.
Sabath, K., Qiu, C., Jonas, S.(2024) Mol Cell 84: 2882-2899.e10
- PubMed: 39032489
- DOI: https://doi.org/10.1016/j.molcel.2024.06.032
- Primary Citation of Related Structures:
8R22, 8R23, 8R2D - PubMed Abstract:
The modular Integrator complex is a transcription regulator that is essential for embryonic development. It attenuates coding gene expression via premature transcription termination and performs 3'-processing of non-coding RNAs. For both activities, Integrator requires endonuclease activity that is harbored by an RNA cleavage module consisting of INTS4-9-11. How correct assembly of Integrator modules is achieved remains unknown. Here, we show that BRAT1 and WDR73 are critical biogenesis factors for the human cleavage module. They maintain INTS9-11 inactive during maturation by physically blocking the endonuclease active site and prevent premature INTS4 association. Furthermore, BRAT1 facilitates import of INTS9-11 into the nucleus, where it is joined by INTS4. Final BRAT1 release requires locking of the mature cleavage module conformation by inositol hexaphosphate (IP 6 ). Our data explain several neurodevelopmental disorders caused by BRAT1, WDR73, and INTS11 mutations as Integrator assembly defects and reveal that IP 6 is an essential co-factor for cleavage module maturation.
- Institute of Molecular Biology and Biophysics, ETH Zurich, Zurich, Switzerland.
Organizational Affiliation: