Fragment-Based Interrogation of the 14-3-3/TAZ Protein-Protein Interaction.
Andlovic, B., Valenti, D., Centorrino, F., Picarazzi, F., Hristeva, S., Hiltmann, M., Wolf, A., Cantrelle, F.X., Mori, M., Landrieu, I., Levy, L.M., Klebl, B., Tzalis, D., Genski, T., Eickhoff, J., Ottmann, C.(2024) Biochemistry 63: 2196-2206
- PubMed: 39172504
- DOI: https://doi.org/10.1021/acs.biochem.4c00248
- Primary Citation of Related Structures:
8R0Z - PubMed Abstract:
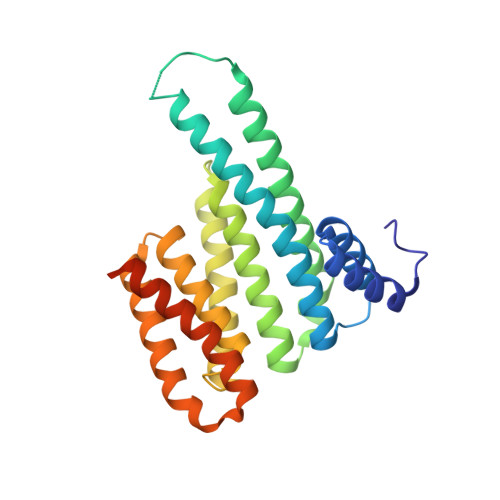
The identification of chemical starting points for the development of molecular glues is challenging. Here, we employed fragment screening and identified an allosteric stabilizer of the complex between 14-3-3 and a TAZ-derived peptide. The fragment binds preferentially to the 14-3-3/TAZ peptide complex and shows moderate stabilization in differential scanning fluorimetry and microscale thermophoresis. The binding site of the fragment was predicted by molecular dynamics calculations to be distant from the 14-3-3/TAZ peptide interface, located between helices 8 and 9 of the 14-3-3 protein. This site was confirmed by nuclear magnetic resonance and X-ray protein crystallography, revealing the first example of an allosteric stabilizer for 14-3-3 protein-protein interactions.
- Lead Discovery Center GmbH, Otto-Hahn-Str. 15, 44227 Dortmund, Germany.
Organizational Affiliation: