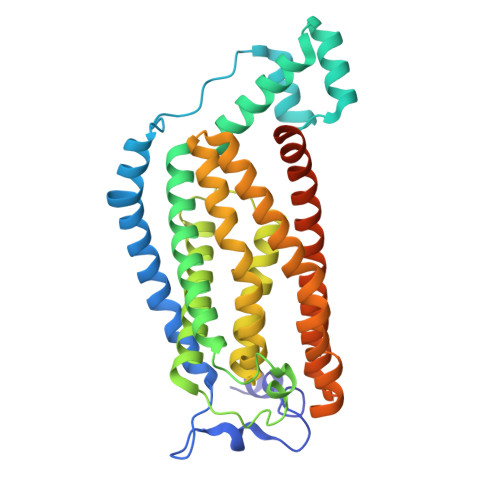
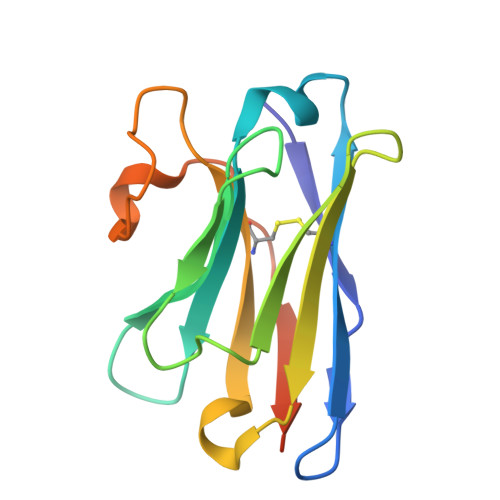
Structural basis of the mechanism and inhibition of a human ceramide synthase.
Pascoa, T.C., Pike, A.C.W., Tautermann, C.S., Chi, G., Traub, M., Quigley, A., Chalk, R., Stefanic, S., Thamm, S., Pautsch, A., Carpenter, E.P., Schnapp, G., Sauer, D.B.(2025) Nat Struct Mol Biol 32: 431-440
- PubMed: 39528795
- DOI: https://doi.org/10.1038/s41594-024-01414-3
- Primary Citation of Related Structures:
8QZ6, 8QZ7, 9EOT - PubMed Abstract:
Ceramides are bioactive sphingolipids crucial for regulating cellular metabolism. Ceramides and dihydroceramides are synthesized by six ceramide synthase (CerS) enzymes, each with specificity for different acyl-CoA substrates. Ceramide with a 16-carbon acyl chain (C16 ceramide) has been implicated in obesity, insulin resistance and liver disease and the C16 ceramide-synthesizing CerS6 is regarded as an attractive drug target for obesity-associated disease. Despite their importance, the molecular mechanism underlying ceramide synthesis by CerS enzymes remains poorly understood. Here we report cryo-electron microscopy structures of human CerS6, capturing covalent intermediate and product-bound states. These structures, along with biochemical characterization, reveal that CerS catalysis proceeds through a ping-pong reaction mechanism involving a covalent acyl-enzyme intermediate. Notably, the product-bound structure was obtained upon reaction with the mycotoxin fumonisin B1, yielding insights into its inhibition of CerS. These results provide a framework for understanding CerS function, selectivity and inhibition and open routes for future drug discovery.
- Centre for Medicines Discovery, Nuffield Department of Medicine, University of Oxford, Oxford, UK. tomas.pascoa@cmd.ox.ac.uk.
Organizational Affiliation: