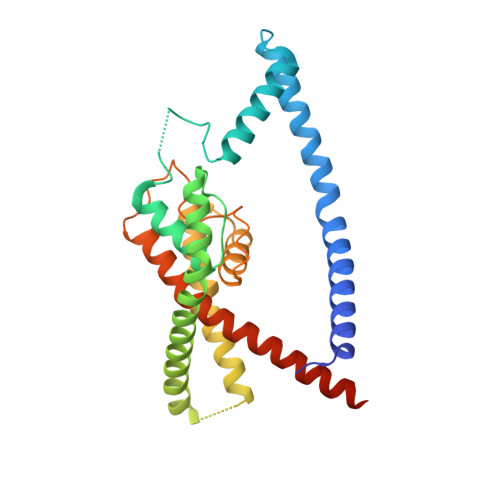
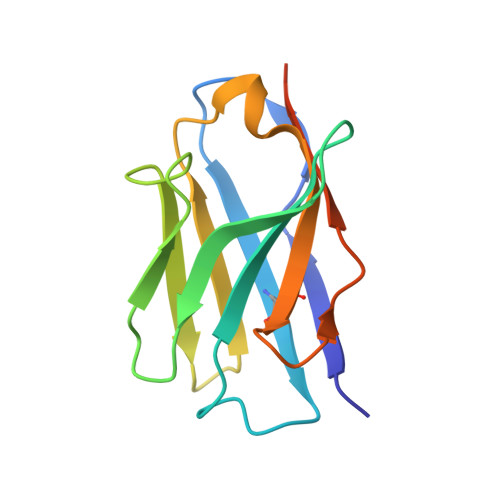
Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Rodstrom, K.E.J., Cloake, A., Sormann, J., Baronina, A., Smith, K.H.M., Pike, A.C.W., Ang, J., Proks, P., Schewe, M., Holland-Kaye, I., Bushell, S.R., Elliott, J., Pardon, E., Baukrowitz, T., Owens, R.J., Newstead, S., Steyaert, J., Carpenter, E.P., Tucker, S.J.(2024) Nat Commun 15: 4173-4173
- PubMed: 38755204
- DOI: https://doi.org/10.1038/s41467-024-48536-2
- Primary Citation of Related Structures:
8QZ1, 8QZ2, 8QZ3, 8QZ4 - PubMed Abstract:
Potassium channels of the Two-Pore Domain (K2P) subfamily, KCNK1-KCNK18, play crucial roles in controlling the electrical activity of many different cell types and represent attractive therapeutic targets. However, the identification of highly selective small molecule drugs against these channels has been challenging due to the high degree of structural and functional conservation that exists not only between K2P channels, but across the whole K + channel superfamily. To address the issue of selectivity, here we generate camelid antibody fragments (nanobodies) against the TREK-2 (KCNK10) K2P K + channel and identify selective binders including several that directly modulate channel activity. X-ray crystallography and CryoEM data of these nanobodies in complex with TREK-2 also reveal insights into their mechanisms of activation and inhibition via binding to the extracellular loops and Cap domain, as well as their suitability for immunodetection. These structures facilitate design of a biparatropic inhibitory nanobody with markedly improved sensitivity. Together, these results provide important insights into TREK channel gating and provide an alternative, more selective approach to modulation of K2P channel activity via their extracellular domains.
- Clarendon Laboratory, Department of Physics, University of Oxford, Oxford, UK.
Organizational Affiliation: