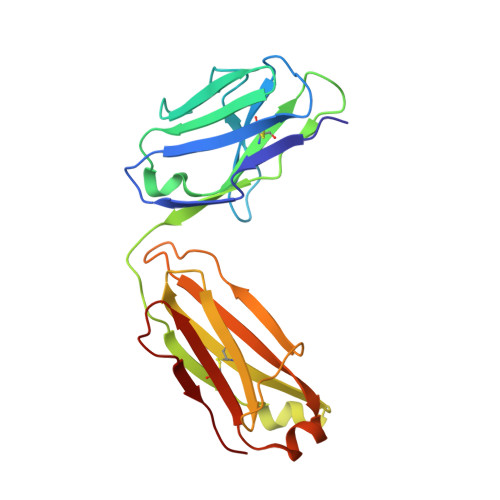
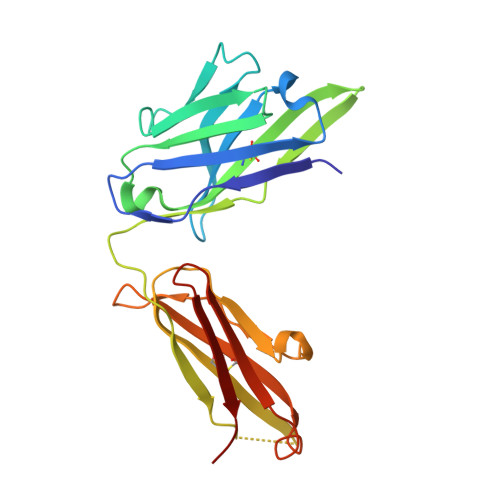
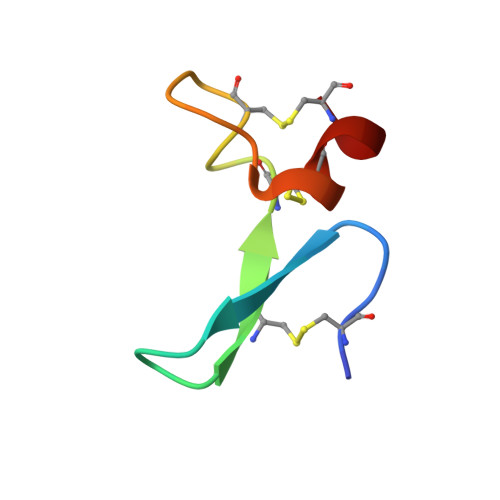
Structure-based humanization of a therapeutic antibody for multiple myeloma.
Marino, S.F., Daumke, O.(2024) J Mol Med (Berl) 102: 1151-1161
- PubMed: 39052065
- DOI: https://doi.org/10.1007/s00109-024-02470-4
- Primary Citation of Related Structures:
8QY9, 8QYA, 8QYB - PubMed Abstract:
The optimal efficacy of xenogeneically generated proteins intended for application in humans requires that their own antigenicity be minimized. This necessary adaptation of antibodies to a humanized version poses challenges since modifications even distant from the binding sites can greatly influence antigen recognition and this is the primary feature that must be maintained during all modifications. Current strategies often rely on grafting and/or randomization/selection to arrive at a humanized variant retaining the binding properties of the original molecule. However, in terms of speed and efficiency, rationally directed approaches can be superior, provided the requisite structural information is available. We present here a humanization procedure based on the high-resolution X-ray structure of a chimaeric IgG against a marker for multiple myeloma. Based on in silico modelling of humanizing amino acid substitutions identified from sequence alignments, we devised a straightforward cloning procedure to rapidly evaluate the proposed sequence changes. Careful inspection of the structure allowed the identification of a potentially problematic amino acid change that indeed disrupted antigen binding. Subsequent optimization of the antigen binding loop sequences resulted in substantial recovery of binding affinity lost in the completely humanized antibody. X-ray structures of the humanized and optimized variants demonstrate that the antigen binding mode is preserved, with surprisingly few direct contacts to antibody atoms. These results underline the importance of structural information for the efficient optimization of protein therapeutics. KEY MESSAGES: Structure-based humanization of an IgG against BCMA, a marker for Multiple Myeloma. Identification of problematic mutations and unexpected modification sites. Structures of the modified IgG-antigen complexes verified predictions. Provision of humanized high-affinity IgGs against BCMA for therapeutic applications.
- Max Delbrück Centrum for Molecular Medicine, Robert-Rössle-Straße 10, 13125, Berlin, Germany. Stephen-Francis.Marino@bfr.bund.de.
Organizational Affiliation: