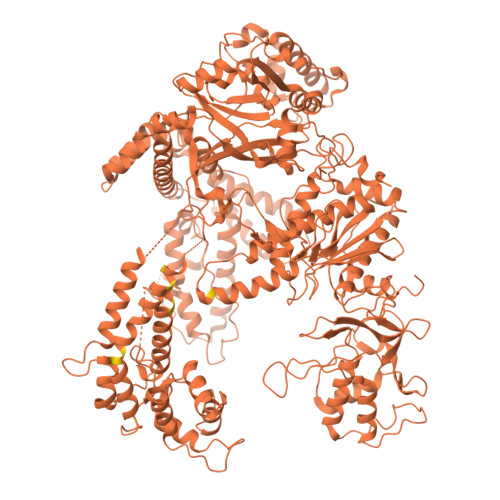
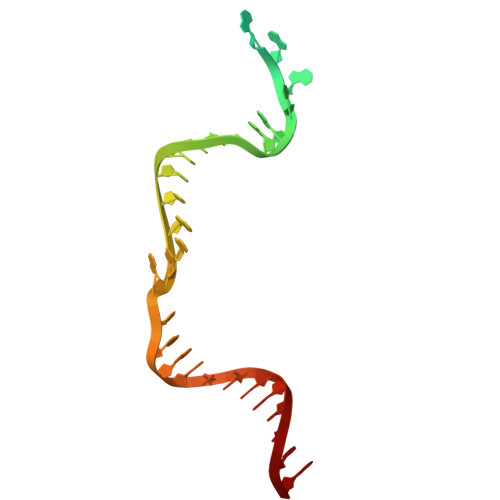
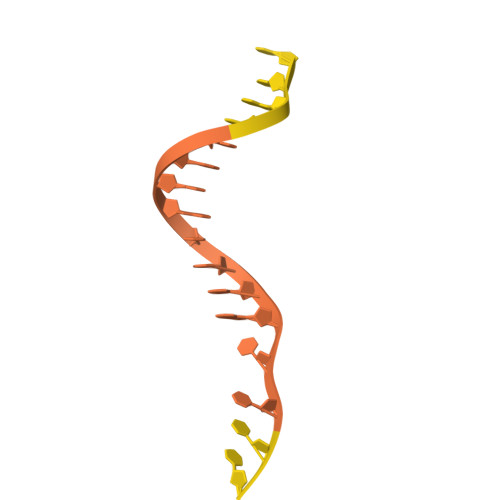
A resurrected ancestor of Cas12a expands target access and substrate recognition for nucleic acid editing and detection.
Jabalera, Y., Tascon, I., Samperio, S., Lopez-Alonso, J.P., Gonzalez-Lopez, M., Aransay, A.M., Abascal-Palacios, G., Beisel, C.L., Ubarretxena-Belandia, I., Perez-Jimenez, R.(2025) Nat Biotechnol 43: 1663-1672
- PubMed: 39482449
- DOI: https://doi.org/10.1038/s41587-024-02461-3
- Primary Citation of Related Structures:
8QWD, 8QWE, 8QWF - PubMed Abstract:
The properties of Cas12a nucleases constrict the range of accessible targets and their applications. In this study, we applied ancestral sequence reconstruction (ASR) to a set of Cas12a orthologs from hydrobacteria to reconstruct a common ancestor, ReChb, characterized by near-PAMless targeting and the recognition of diverse nucleic acid activators and collateral substrates. ReChb shares 53% sequence identity with the closest Cas12a ortholog but no longer requires a T-rich PAM and can achieve genome editing in human cells at sites inaccessible to the natural FnCas12a or the engineered and PAM-flexible enAsCas12a. Furthermore, ReChb can be triggered not only by double-stranded DNA but also by single-stranded RNA and DNA targets, leading to non-specific collateral cleavage of all three nucleic acid substrates with similar efficiencies. Finally, tertiary and quaternary structures of ReChb obtained by cryogenic electron microscopy reveal the molecular details underlying its expanded biophysical activities. Overall, ReChb expands the application space of Cas12a nucleases and underscores the potential of ASR for enhancing CRISPR technologies.
- Center for Cooperative Research in Biosciences (CIC bioGUNE), Basque Research and Technology Alliance (BRTA), Derio, Spain.
Organizational Affiliation: