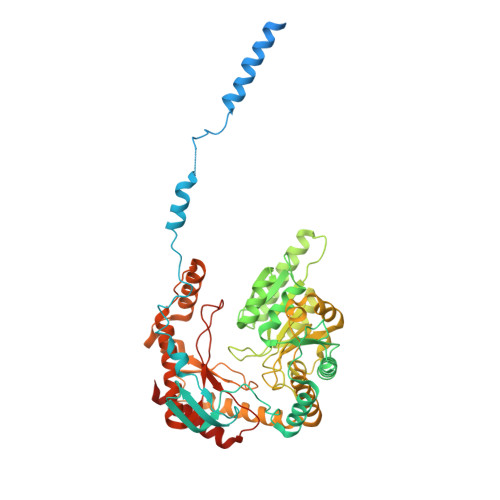
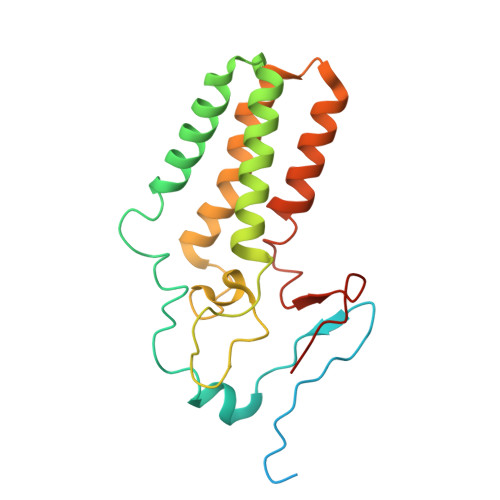
The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins.
Korner, C., Schafer, J.H., Esch, B.M., Parey, K., Walter, S., Teis, D., Januliene, D., Schmidt, O., Moeller, A., Frohlich, F.(2024) Cell Rep 43: 114627-114627
- PubMed: 39167489
- DOI: https://doi.org/10.1016/j.celrep.2024.114627
- Primary Citation of Related Structures:
8QOF, 8QOG - PubMed Abstract:
Sphingolipid levels are crucial determinants of neurodegenerative disorders and therefore require tight regulation. The Orm protein family and ceramides inhibit the rate-limiting step of sphingolipid biosynthesis-the condensation of L-serine and palmitoyl-coenzyme A (CoA). The yeast isoforms Orm1 and Orm2 form a complex with the serine palmitoyltransferase (SPT). While Orm1 and Orm2 have highly similar sequences, they are differentially regulated, though the mechanistic details remain elusive. Here, we determine the cryoelectron microscopy structure of the SPT complex containing Orm2. Complementary in vitro activity assays and genetic experiments with targeted lipidomics demonstrate a lower activity of the SPT-Orm2 complex than the SPT-Orm1 complex. Our results suggest a higher inhibitory potential of Orm2, despite the similar structures of the Orm1- and Orm2-containing complexes. The high conservation of SPT from yeast to man implies different regulatory capacities for the three human ORMDL isoforms, which might be key for understanding their role in sphingolipid-mediated neurodegenerative disorders.
- Bioanalytical Chemistry Section, Department of Biology/Chemistry, Osnabrück University, 49076 Osnabrück, Germany.
Organizational Affiliation: