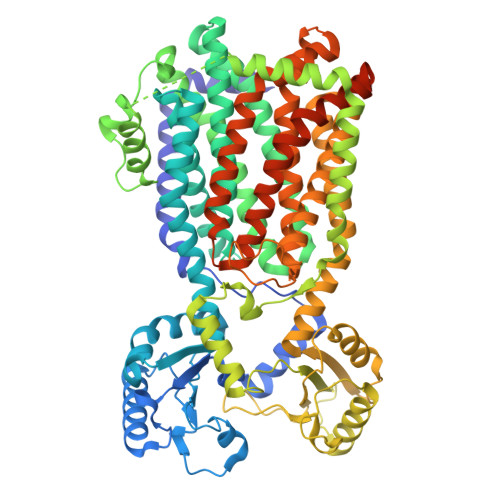
Cryo-EM structure of the trehalose monomycolate transporter, MmpL3, reconstituted into peptidiscs.
Couston, J., Guo, Z., Wang, K., Gourdon, P., Blaise, M.(2023) Curr Res Struct Biol 6: 100109-100109
- PubMed: 38034087
- DOI: https://doi.org/10.1016/j.crstbi.2023.100109
- Primary Citation of Related Structures:
8QKK - PubMed Abstract:
Mycobacteria have an atypical thick and waxy cell wall. One of the major building blocks of such mycomembrane is trehalose monomycolate (TMM). TMM is a mycolic acid ester of trehalose that possesses long acyl chains with up to 90 carbon atoms. TMM represents an essential component of mycobacteria and is synthesized in the cytoplasm, and then flipped over the plasma membrane by a specific transporter known as MmpL3. Over the last decade, MmpL3 has emerged as an attractive drug target to combat mycobacterial infections. Recent three-dimensional structures of MmpL3 determined by X-ray crystallography and cryo-EM have increased our understanding of the TMM transport, and the mode of action of inhibiting compounds. These structures were obtained in the presence of detergent and/or in a lipidic environment. In this study, we demonstrate the possibility of obtaining a high-quality cryo-EM structure of MmpL3 without any presence of detergent through the reconstitution of the protein into peptidiscs. The structure was determined at an overall resolution of 3.2 Å and demonstrates that the overall structure of MmpL3 is preserved as compared to previous structures. Further, the study identified a new structural arrangement of the linker that fuses the two subdomains of the transmembrane domain, suggesting the feature may serve a role in the transport process.
- IRIM, CNRS, University of Montpellier, Montpellier, France.
Organizational Affiliation: