Enhancing the Features of DNA Mimic Foldamers for Structural Investigations.
Corvaglia, V., Wu, J., Deepak, D., Loos, M., Huc, I.(2024) Chemistry 30: e202303650-e202303650
- PubMed: 38193643
- DOI: https://doi.org/10.1002/chem.202303650
- Primary Citation of Related Structures:
8QHM - PubMed Abstract:
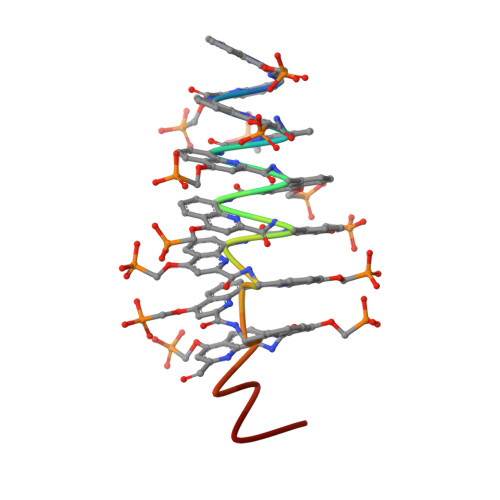
DNA mimic foldamers based on aromatic oligoamide helices bearing anionic phosphonate side chains have been shown to bind to DNA-binding proteins sometimes orders of magnitude better than DNA itself. Here, we introduce new features in the DNA mimic foldamers to facilitate structural investigations of their interactions with proteins. Thirteen new foldamer sequences have been synthesized and characterized using NMR, circular dichroism, molecular modeling, and X-ray crystallography. The results show that foldamer helix handedness can be quantitatively biased by means of a single stereogenic center, that the foldamer structure can be made C 2 -symmetrical as in palindromic B-DNA sequences, and that associations between foldamer helices can be promoted utilizing dedicated C-terminal residues that act as sticky ends in B-DNA structures.
Organizational Affiliation:
Department of Pharmacy, Ludwig-Maximilians-Universität München, Butenandtstr. 5-13, 81377, Munich, Germany.